| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,822,086 – 11,822,204 |
| Length | 118 |
| Max. P | 0.873693 |

| Location | 11,822,086 – 11,822,179 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -22.98 |
| Energy contribution | -24.87 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11822086 93 + 20766785 GUGUCAAUUGCAGCGCCACCACUGCCAUCACAAUGAGGGACCAAUGACCAGUAGU--GUCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGU-GU-------CU ............((((..(((((((((((.((......(((((....((((....--..))))...)))))..)))))))))))))...))-))-------.. ( -36.40) >DroSec_CAF1 12642 93 + 1 GUGUCAAUUGCAGCGCCACCACUGCCAUCACAAUGAGGGACCAAUGACCAGUAGU--GGCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGU-GU-------CU ............((((..(((((((((((.((......(((((....(((((...--.)))))...)))))..)))))))))))))...))-))-------.. ( -37.80) >DroSim_CAF1 11502 93 + 1 GUGUCAAUUGCAGCGCCACCACUGCCAUCACAAUGAGGGACCAAUGACCAGUAGU--GGCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGU-GU-------CU ............((((..(((((((((((.((......(((((....(((((...--.)))))...)))))..)))))))))))))...))-))-------.. ( -37.80) >DroEre_CAF1 12414 86 + 1 GUGUCAAUUGCAGCGCCACCACUGCCAUCACAAUGAGGGACCAAUGACCAGUAGU--GGCUGGAAGU-------GGAUGGCAGUGGAUAGU-GU-------CU ............((((..(((((((((((.((.((......)).)).(((((...--.)))))....-------.)))))))))))...))-))-------.. ( -32.00) >DroYak_CAF1 12599 95 + 1 GUGUCAAUUGCAGCGCCACCACUGCCAUCACAAUGAGGGACCAAUGACCAGUAGUGCGGCUGGAAGU-------GGAUGGCAGUGGAUAGU-GUCUUAUGUCU ............((((..(((((((((((.((.((......)).)).(((((......)))))....-------.)))))))))))...))-))......... ( -32.20) >DroAna_CAF1 11673 74 + 1 GUGUCAAUUGCAGCGCCAUCACCAACAUCACAAUGAGGGACCAAUGACCAGUCCC--AGC--------------------CACUAAACACUAGC-------CU ((((.....((..............(((....))).(((((.........)))))--.))--------------------......))))....-------.. ( -11.70) >consensus GUGUCAAUUGCAGCGCCACCACUGCCAUCACAAUGAGGGACCAAUGACCAGUAGU__GGCUGGAAGU_______GGAUGGCAGUGGAUAGU_GU_______CU ((((........))))..(((((((((((.((.((......)).)).(((((......)))))............)))))))))))................. (-22.98 = -24.87 + 1.89)

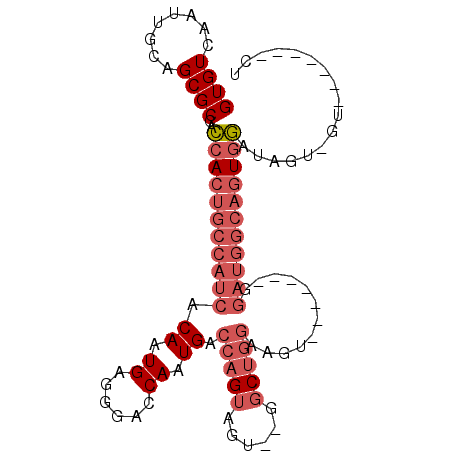
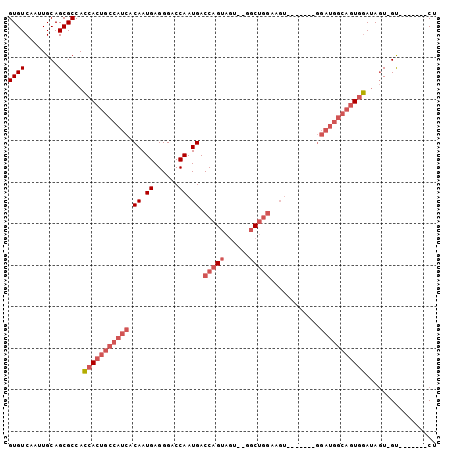
| Location | 11,822,086 – 11,822,179 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -19.18 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11822086 93 - 20766785 AG-------AC-ACUAUCCACUGCCAUCCACUAACCACUUCCAGAC--ACUACUGGUCAUUGGUCCCUCAUUGUGAUGGCAGUGGUGGCGCUGCAAUUGACAC ((-------.(-.(((.(((((((((((((...((((...((((..--....))))....)))).......)).)))))))))))))).)))........... ( -31.70) >DroSec_CAF1 12642 93 - 1 AG-------AC-ACUAUCCACUGCCAUCCACUAACCACUUCCAGCC--ACUACUGGUCAUUGGUCCCUCAUUGUGAUGGCAGUGGUGGCGCUGCAAUUGACAC ((-------.(-.(((.(((((((((((((...((((...((((..--....))))....)))).......)).)))))))))))))).)))........... ( -31.70) >DroSim_CAF1 11502 93 - 1 AG-------AC-ACUAUCCACUGCCAUCCACUAACCACUUCCAGCC--ACUACUGGUCAUUGGUCCCUCAUUGUGAUGGCAGUGGUGGCGCUGCAAUUGACAC ((-------.(-.(((.(((((((((((((...((((...((((..--....))))....)))).......)).)))))))))))))).)))........... ( -31.70) >DroEre_CAF1 12414 86 - 1 AG-------AC-ACUAUCCACUGCCAUCC-------ACUUCCAGCC--ACUACUGGUCAUUGGUCCCUCAUUGUGAUGGCAGUGGUGGCGCUGCAAUUGACAC ((-------.(-.(((.((((((((((((-------(...((((((--(....))))...)))........)).)))))))))))))).)))........... ( -28.10) >DroYak_CAF1 12599 95 - 1 AGACAUAAGAC-ACUAUCCACUGCCAUCC-------ACUUCCAGCCGCACUACUGGUCAUUGGUCCCUCAUUGUGAUGGCAGUGGUGGCGCUGCAAUUGACAC .......((.(-.(((.(((((((((((.-------....((((........)))).((.(((....))).)).)))))))))))))).)))........... ( -28.00) >DroAna_CAF1 11673 74 - 1 AG-------GCUAGUGUUUAGUG--------------------GCU--GGGACUGGUCAUUGGUCCCUCAUUGUGAUGUUGGUGAUGGCGCUGCAAUUGACAC ..-------....(((((..(..--------------------((.--(((((..(...)..)))))((((((......))))))....))..)....))))) ( -21.40) >consensus AG_______AC_ACUAUCCACUGCCAUCC_______ACUUCCAGCC__ACUACUGGUCAUUGGUCCCUCAUUGUGAUGGCAGUGGUGGCGCUGCAAUUGACAC .............(((.(((((((((((............((((........)))).((.(((....))).)).))))))))))))))............... (-19.18 = -20.02 + 0.84)

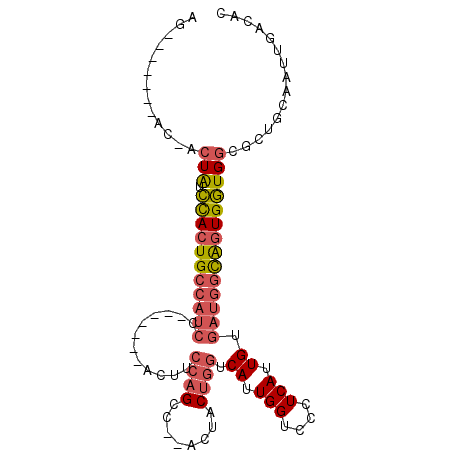
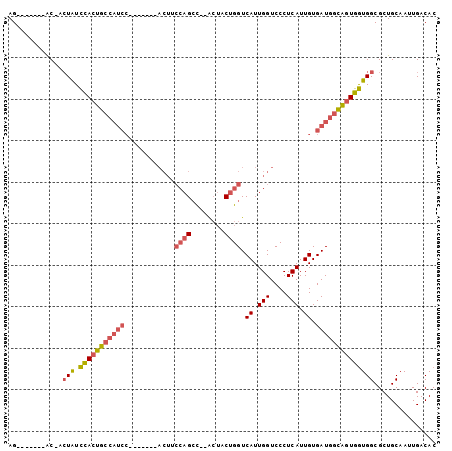
| Location | 11,822,113 – 11,822,204 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.27 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11822113 91 + 20766785 UCACAAUGAGGGACCAAUGACCAGUAGU--GUCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGUGU-------CUAAUGUCUAAUGGCCAGUGGCCAAUU ...........(((((....((((....--..))))...)))))..(((.((((..((((((....-------....))))))...))))....)))... ( -26.30) >DroSec_CAF1 12669 91 + 1 UCACAAUGAGGGACCAAUGACCAGUAGU--GGCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGUGU-------CUAAUGUCUAAUGGCCAGUGGCCAAUU ...........(((((....(((((...--.)))))...)))))..(((.((((..((((((....-------....))))))...))))....)))... ( -27.70) >DroSim_CAF1 11529 91 + 1 UCACAAUGAGGGACCAAUGACCAGUAGU--GGCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGUGU-------CUAAUGUCUAAUGGCCAGUGGCCAAUU ...........(((((....(((((...--.)))))...)))))..(((.((((..((((((....-------....))))))...))))....)))... ( -27.70) >DroEre_CAF1 12441 84 + 1 UCACAAUGAGGGACCAAUGACCAGUAGU--GGCUGGAAGU-------GGAUGGCAGUGGAUAGUGU-------CUAAUGUCUAAUGGCCAGUGGCCAAUU ((((.........(((....(((((...--.)))))...)-------)).((((..((((((....-------....))))))...))))))))...... ( -24.80) >DroYak_CAF1 12626 93 + 1 UCACAAUGAGGGACCAAUGACCAGUAGUGCGGCUGGAAGU-------GGAUGGCAGUGGAUAGUGUCUUAUGUCUAAUGUCUAAUGGCCAGUGGCCAAUU .............(((....(((((......)))))...)-------))..((((.((((((........)))))).))))...(((((...)))))... ( -25.90) >consensus UCACAAUGAGGGACCAAUGACCAGUAGU__GGCUGGAAGUGGUUAGUGGAUGGCAGUGGAUAGUGU_______CUAAUGUCUAAUGGCCAGUGGCCAAUU ..........((.(((....(((((......)))))..............((((..((((((...............))))))...)))).))))).... (-22.54 = -22.74 + 0.20)

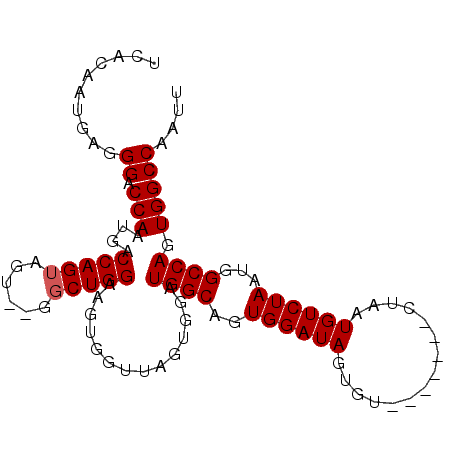
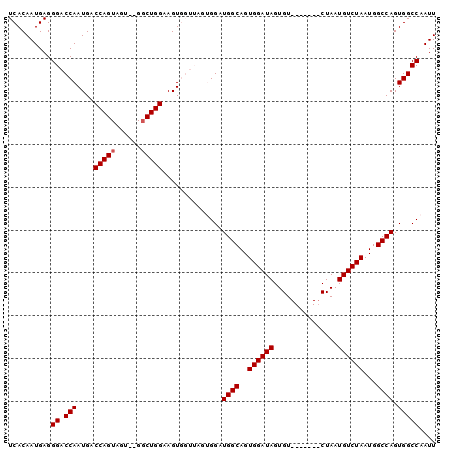
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:53 2006