
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,820,450 – 11,820,553 |
| Length | 103 |
| Max. P | 0.776644 |

| Location | 11,820,450 – 11,820,553 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -16.68 |
| Energy contribution | -17.90 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11820450 103 - 20766785 --AAAGUAACUAAACGCAGAGUUCCAC-------------AAAAUAUUGUUGCCUGCGAUCGUGGAUCGUGGCAUGCGCACUCUGAUCGAGAUCGCAGCGACAGAUAGCGGAGCAAAG --..................(((((.(-------------......(((((((.(((((((.(.(((((..((....))....))))).)))))))))))))))...).))))).... ( -32.90) >DroPse_CAF1 9944 112 - 1 --A-AGAAA---AAAACAGAACUGCACGAUAAAUAAACAGAAAAUAUUGUUGCAAGAGAUCGUGGAUCGUUGCAUGCGCACUCUGAUCGAGAUCUCAGCGACAGAUAGCGGACCCAAG --.-.....---.........((((.(............)......(((((((..((((((.(.(((((.(((....)))...))))).))))))).)))))))...))))....... ( -26.40) >DroSec_CAF1 10972 102 - 1 --AAAGUAACUA-GCGCAGAGUUCCAC-------------AAAAUAUUGUUGCCUGCGAUCGUGGAUAGUGGCAUGCGCACUCUGAUCGAGAUCGCAGCGACAGAUAGCGGAGCAAAG --..........-.......(((((.(-------------......(((((((.(((((((.(((.(((.(((....)).).))).))).))))))))))))))...).))))).... ( -31.90) >DroEre_CAF1 10753 102 - 1 --A-AGUAACUAAACGCAGAGUUCCAC-------------AAAAUAUUGUUGCCAGCGAUCGUGGAUCGUGGCAUGCGCACUCUGAUCGAGAUCGCAGCGACAGAUAGCGGAGCAAAG --.-................(((((.(-------------......(((((((..((((((.(.(((((..((....))....))))).))))))).)))))))...).))))).... ( -30.40) >DroAna_CAF1 10203 93 - 1 AGAAAAACACUAAACGCAGAGUUACAA-------------AAAAUAUGGUUGGCUGCGAUCGUGAAUCGUGGCAUGC-----------GAGAUCGCAGCGACAGAUAGCG-ACCAAAG ..............(((((.....((.-------------......)).....))))).((((..(((((.((.(((-----------(....)))))).)).))).)))-)...... ( -24.30) >DroPer_CAF1 10733 113 - 1 --A-AGAAAA--AAAACAGAACUGCACGAUAAAUAAACAGAAAAUAUUGUUGCAAGAGAUCGUGGAUCGUUGCAUGCGCACUCUGAUCGAGAUCUCAGCGACAGAUAGCGGACCCAAG --.-......--.........((((.(............)......(((((((..((((((.(.(((((.(((....)))...))))).))))))).)))))))...))))....... ( -26.40) >consensus __A_AGAAACUAAACGCAGAGUUCCAC_____________AAAAUAUUGUUGCCAGCGAUCGUGGAUCGUGGCAUGCGCACUCUGAUCGAGAUCGCAGCGACAGAUAGCGGACCAAAG ..............................................(((((((..((((((.(.(((((..((....))....))))).))))))).))))))).............. (-16.68 = -17.90 + 1.22)

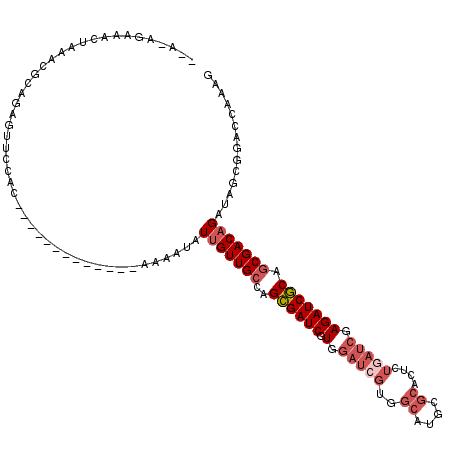
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:51 2006