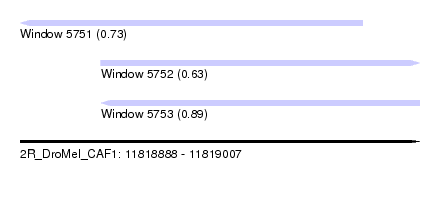
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,818,888 – 11,819,007 |
| Length | 119 |
| Max. P | 0.893593 |

| Location | 11,818,888 – 11,818,990 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -25.08 |
| Energy contribution | -26.44 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
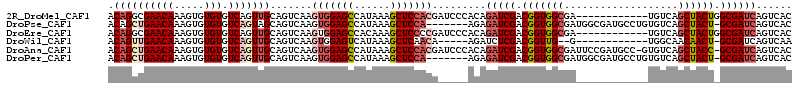
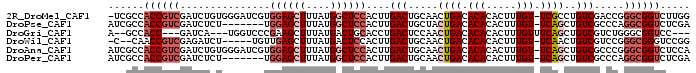
>2R_DroMel_CAF1 11818888 102 - 20766785 ACAGGCGAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGA------------UGUCAGCUACUGGCGAUCAGUCAC ...(((.......(((.(..((..(((....)))(((((((......)))))))))..)))).(((((.(((((((..------------.....))))))).))))).))).. ( -38.90) >DroPse_CAF1 8421 106 - 1 ACAGCUGAACAAAGUGUGUGUCAGUAGCAGUCAAGUGGAGCCAUAAAGCUCCA-------AGAGAUCGACGGUGGCGAUGGCGAUGCCUGUGUCAGCUACU-GCGAUCAGUCAC ...((((((((.....))).))))).....((...((((((......))))))-------.))(((((.(((((((((((((...)))...))).))))))-))))))...... ( -36.90) >DroEre_CAF1 9224 102 - 1 ACAGGCGAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCACAAAGCUCCCCGAUCCCACAGAUCGACGGUGGCGA------------UGUCAGCUACUGGCGAUCAGUCAC ...(((.......(((.(..((..(((....)))(.(((((......))))).)))..)))).(((((.(((((((..------------.....))))))).))))).))).. ( -34.10) >DroWil_CAF1 14584 94 - 1 ACAGUUGAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGUCAUAAAGCUCAACA-----AGAUCUCGACGGUUG--G------------UGGCAACAACU-GCGAUCAGUCAA (((.((.....)).)))......((((..(((..((.((((......)))).)).-----.)))..))))(..((--(------------(.(((.....)-)).))))..).. ( -22.80) >DroAna_CAF1 8787 112 - 1 ACAGCUGAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGAUUCCGAUGCC-GUGUCAGCUACC-GCGAUCAGUCAC .((((((((((.....))).))))))).......(((((((......))))))).........(((((.((((((((((..((....)-).))).))))))-))))))...... ( -44.20) >DroPer_CAF1 9206 106 - 1 ACAGCUGAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCA-------AGAGAUCGACGGUGGCGAUGGCGAUGCCUGUGUCAGCUACU-GCGAUCAGUCAC .((((((((((.....))).)))))))...((...((((((......))))))-------.))(((((.(((((((((((((...)))...))).))))))-))))))...... ( -40.00) >consensus ACAGCUGAACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACG_____ACAGAUCGACGGUGGCGA____________UGUCAGCUACU_GCGAUCAGUCAC .((((((((((.....))).))))))).......(((((((......))))))).........(((((.(((((((...................)))))).))))))...... (-25.08 = -26.44 + 1.36)
| Location | 11,818,912 – 11,819,007 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.63 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
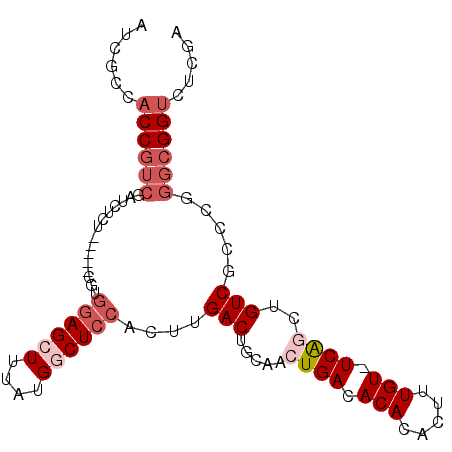
>2R_DroMel_CAF1 11818912 95 + 20766785 -UCGCCACCGUCGAUCUGUGGGAUCGUGGAGCUUUAUGGCUCCACUUGACUGCAACUGACACACACUUUGU-UCGCCUGUCGACCGGGCGGUCUUGG -(((((.(.(((((((.....))))((((((((....))))))))..(((.(((((.............))-).))..)))))).))))))...... ( -34.82) >DroPse_CAF1 8455 89 + 1 AUCGCCACCGUCGAUCUCU-------UGGAGCUUUAUGGCUCCACUUGACUGCUACUGACACACACUUUGU-UCAGCUGUCGCCCAGGCGGUCUCGA .(((..((((((.......-------(((((((....)))))))...(((.(((...((((.......)))-).))).))).....))))))..))) ( -29.90) >DroGri_CAF1 24490 86 + 1 A--GCCACC---GAUCA---UGGUCCCGAAGCUUUAUGACUGCACCUGACUCCAACUGACACACACUUUGUUUCAGCUGUCGUCUGGGCGGUCC--- (--((....---(((..---..))).....)))....((((((.((.(((..((.((((.(((.....))).)))).))..))).)))))))).--- ( -26.00) >DroWil_CAF1 14607 88 + 1 -C--CAACCGUCGAGAUCU-----UGUUGAGCUUUAUGACUCCACUUGACUGCAACUGACACACACUUUGU-UCAACUGUCGUCCGGGCGGGUCCGG -.--...(((..(((.((.-----....)).)))...(((((..((.(((.(((..(((.(((.....)))-)))..))).))).))..)))))))) ( -18.90) >DroAna_CAF1 8820 96 + 1 AUCGCCACCGUCGAUCUGUGGGAUCGUGGAGCUUUAUGGCUCCACUUGACUGCAACUGACACACACUUUGU-UCAGCUGUCGCCCGGGCGGUCUCCA ((((((...(((((((.....))))((((((((....))))))))..))).(((.((((.(((.....)))-)))).)))......))))))..... ( -36.20) >DroPer_CAF1 9240 89 + 1 AUCGCCACCGUCGAUCUCU-------UGGAGCUUUAUGGCUCCACUUGACUGCAACUGACACACACUUUGU-UCAGCUGUCGCCCAGGCGGUCUCGA .(((..((((((..((...-------(((((((....)))))))...))..(((.((((.(((.....)))-)))).)))......))))))..))) ( -29.10) >consensus AUCGCCACCGUCGAUCUCU_____CGUGGAGCUUUAUGGCUCCACUUGACUGCAACUGACACACACUUUGU_UCAGCUGUCGCCCGGGCGGUCUCGA ......((((((...............((((((....))))))....(((.....((((.(((.....))).))))..))).....))))))..... (-18.10 = -19.63 + 1.53)
| Location | 11,818,912 – 11,819,007 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -25.55 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
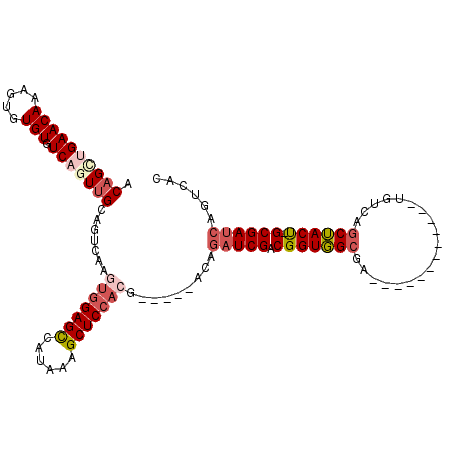
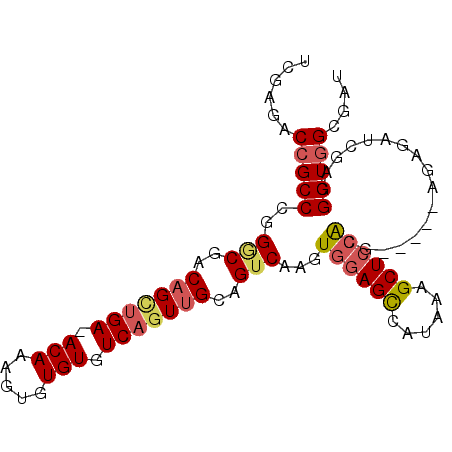
>2R_DroMel_CAF1 11818912 95 - 20766785 CCAAGACCGCCCGGUCGACAGGCGA-ACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGA- ....((((....))))((((.(((.-......))).)))).((..(.(((..(((((((......)))))))((((.....))))))).)..))..- ( -39.80) >DroPse_CAF1 8455 89 - 1 UCGAGACCGCCUGGGCGACAGCUGA-ACAAAGUGUGUGUCAGUAGCAGUCAAGUGGAGCCAUAAAGCUCCA-------AGAGAUCGACGGUGGCGAU (((..((((.(..(((..(.(((((-(((.....))).))))).)..)))...((((((......))))))-------.......).))))..))). ( -28.80) >DroGri_CAF1 24490 86 - 1 ---GGACCGCCCAGACGACAGCUGAAACAAAGUGUGUGUCAGUUGGAGUCAGGUGCAGUCAUAAAGCUUCGGGACCA---UGAUC---GGUGGC--U ---.(((.((((.(((..(((((((.(((.....))).)))))))..))).)).)).)))....((((((((.....---...))---)).)))--) ( -31.20) >DroWil_CAF1 14607 88 - 1 CCGGACCCGCCCGGACGACAGUUGA-ACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGUCAUAAAGCUCAACA-----AGAUCUCGACGGUUG--G- ((((((((((...(((..((..(((-(((.....))).)))..))..)))..)))).)))....((.((....-----.)).))...)))...--.- ( -22.30) >DroAna_CAF1 8820 96 - 1 UGGAGACCGCCCGGGCGACAGCUGA-ACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACGAUCCCACAGAUCGACGGUGGCGAU ....(.(((((..(((..(((((((-(((.....))).)))))))..)))..(((((((......)))))))((((.....))))...))))))... ( -38.40) >DroPer_CAF1 9240 89 - 1 UCGAGACCGCCUGGGCGACAGCUGA-ACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCA-------AGAGAUCGACGGUGGCGAU (((..((((.(..(((..(((((((-(((.....))).)))))))..)))...((((((......))))))-------.......).))))..))). ( -32.60) >consensus UCGAGACCGCCCGGGCGACAGCUGA_ACAAAGUGUGUGUCAGUUGCAGUCAAGUGGAGCCAUAAAGCUCCACG_____AGAGAUCGACGGUGGCGAU ......(((((..(((..(((((((.(((.....))).)))))))..)))...((((((......)))))).................))))).... (-25.55 = -26.08 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:50 2006