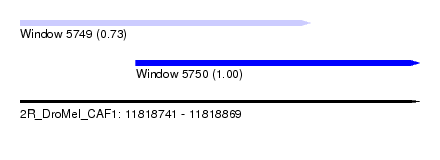
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,818,741 – 11,818,869 |
| Length | 128 |
| Max. P | 0.999502 |

| Location | 11,818,741 – 11,818,834 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
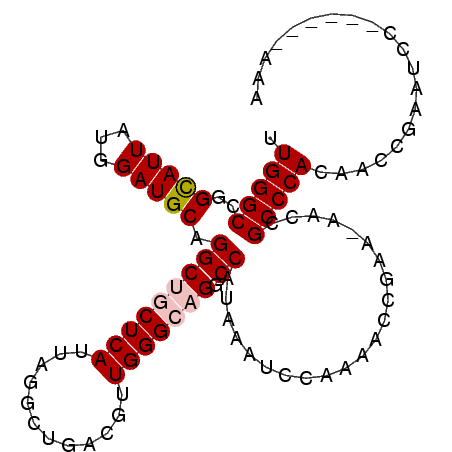
>2R_DroMel_CAF1 11818741 93 + 20766785 UUGGGCCGGCAUUAUGGAUGCAGGCUGCUCAUUAGGCUGACGUUGGGCAGGCCAUAAAUCCAAAAGCGAA-AACCGCCCACAACCGAAUCC------AAA (((((.(((.....(((((...(((((((((...(.....)..)))))).)))....)))))...(((..-...)))......)))..)))------)). ( -28.70) >DroPse_CAF1 8301 76 + 1 UUGGGCCGGUAUUAUGGAUGCAGGCAACUCAUUAAACUGACGCUGGGCAGGCCAUAAAUCC--------------GCCCACC--CG--UCC------AUA .(((((.((..((((((.(((.(((...(((......))).)))..)))..))))))..))--------------)))))..--..--...------... ( -25.40) >DroSec_CAF1 9243 100 + 1 UUGGGCCGGCAUUAUGGAUGCAGGCUGCUCAUUAGGCUGACGUUGGGCAGGCCAUAAAUCCAAAACCGAACAACCGCCCACAACCGAAUCCAAAUCCAAA .((((((((.....(((((...(((((((((...(.....)..)))))).)))....)))))...))).......))))).................... ( -27.81) >DroEre_CAF1 9083 93 + 1 UUGGGCCGGCAUUAUGGAUGCAGGCUGCUCAUUAGGCUGACGUUGGGCAGCCCAUAAAUCCAAAACCGAA-AACCGCCCACAACCGAAUCC------AAA .((((((((.....(((((...(((((((((...(.....)..))))))))).....)))))...)))..-....)))))...........------... ( -31.20) >DroYak_CAF1 9165 93 + 1 UUGGGCCGGCAUUAUGGAUGCAGGCUGCUCAUUAGGCUGACGUUGGGCAGGCCAUAAAUCCAAAACCGAA-AACCGCCCACAACCGAAUCC------AAA .((((((((.....(((((...(((((((((...(.....)..)))))).)))....)))))...)))..-....)))))...........------... ( -28.00) >DroPer_CAF1 9086 76 + 1 UUGGGCCGGUAUUAUGGAUGCAGGCAACUCAUUAAACUGACGCUGGGCAGGCCAUAAAUCC--------------GCCCACC--CG--UCC------AUA .(((((.((..((((((.(((.(((...(((......))).)))..)))..))))))..))--------------)))))..--..--...------... ( -25.40) >consensus UUGGGCCGGCAUUAUGGAUGCAGGCUGCUCAUUAGGCUGACGUUGGGCAGGCCAUAAAUCCAAAACCGAA_AACCGCCCACAACCGAAUCC______AAA .(((((..(((((...))))).(((((((((............))))))).))......................))))).................... (-20.69 = -21.13 + 0.44)

| Location | 11,818,778 – 11,818,869 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -8.28 |
| Consensus MFE | -7.97 |
| Energy contribution | -7.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
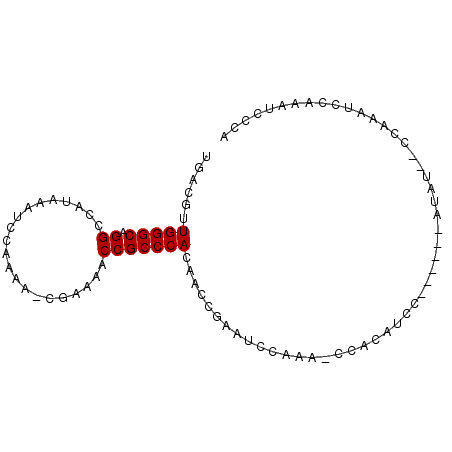
>2R_DroMel_CAF1 11818778 91 + 20766785 UGACGUUGGGCAGGCCAUAAAUCCAAAAGCGAAAACCGCCCACAACCGAAUCCAAAACCACAUCCAUAUCCAUAU--CCAAAUCCAAAUCCCA ....((.((((.((.....................))))))))................................--................ ( -8.20) >DroWil_CAF1 14396 73 + 1 UGACGCUGGGCAGGCCAUAAAUCCAUU-----CAACCGCCCAC---CCAUUACAA------AUUC------AAAAAGAAAAAACCAAUUCCCA ......((((..(((............-----.....)))..)---)))......------....------...................... ( -8.43) >DroYak_CAF1 9202 85 + 1 UGACGUUGGGCAGGCCAUAAAUCCAAAACCGAAAACCGCCCACAACCGAAUCCAAAUCCACAUCC------AUAU--CCAUAUCCAAAUCCCA ....((.((((.((.....................))))))))......................------....--................ ( -8.20) >consensus UGACGUUGGGCAGGCCAUAAAUCCAAAA_CGAAAACCGCCCACAACCGAAUCCAAA_CCACAUCC______AUAU__CCAAAUCCAAAUCCCA ......(((((.((.....................)))))))................................................... ( -7.97 = -7.97 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:47 2006