| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,818,024 – 11,818,117 |
| Length | 93 |
| Max. P | 0.990426 |

| Location | 11,818,024 – 11,818,117 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.94 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.73 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
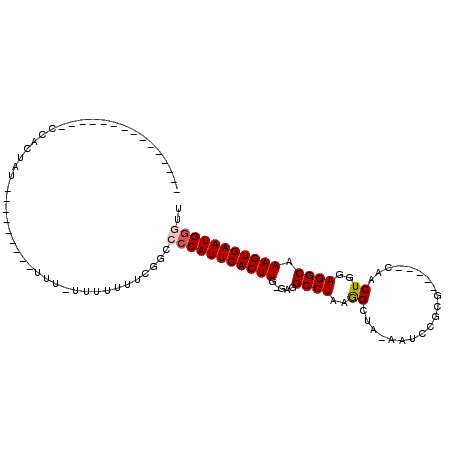
>2R_DroMel_CAF1 11818024 93 + 20766785 -----------UUCUCCUCUAUGUUUGUUUUUUC-UUUUUUUCGGUCCCAUUCACUUG-GAGGCCUAAGGCUA-AAUCGGCG-----CAACUGGAGGCAAAGUGAAUGGUUU -----------.......................-............((((((((((.-...((((.(((((.-....))).-----...))..)))).))))))))))... ( -21.80) >DroPse_CAF1 7462 84 + 1 ---------------CCACAGUG------------UGAGUGUGUGCCACAUUCACUUGUGGGGCCUAAGGCCUAAGAUCGCGACUGGAGACUGGAGGCAAAGUGAAUGGAA- ---------------((((((.(------------((((((((...)))))))))))))))((((...)))).......((..(((....).))..)).............- ( -28.60) >DroSec_CAF1 8550 72 + 1 --------------------------------UU-UCUUUUUCGGCCCCAUUCACUUG-GAGGCCUAAAGCUA-AAUCGGCG-----CAACUGGAGGCAAAGUGAAUGGGUU --------------------------------..-...........(((((((((((.-...((((...(((.-....))).-----.......)))).))))))))))).. ( -24.90) >DroSim_CAF1 8468 73 + 1 -------------------------------UUU-UCUUUUUCGGCCCCAUUCACUUG-GAGGCCUAAGGCUA-AAUCGGCG-----CAACUGGAGGCAAAGUGAAUGGGUU -------------------------------...-...........(((((((((((.-...((((.(((((.-....))).-----...))..)))).))))))))))).. ( -25.10) >DroEre_CAF1 8392 85 + 1 -----------GGCUCCACUAU-------U-UUU-UUCUUUUGGGCCCCAUUCACUUG-GGGGCCUAAGGCUA-AAGCCGCG-----CAACUGGAGGCAAAGUGAAUGGGUU -----------(((.(((....-------.-...-......))))))((((((((((.-...((((..(((..-..)))...-----.......)))).))))))))))... ( -30.04) >DroYak_CAF1 8419 97 + 1 UUUUUUUUUUGUGCAACACUAU-------U-UUUACUCUGUUGGGCCCCAUUCACUUG-GGGGCCUAAGGCUA-AAGCCGCG-----CCACUGGAGGCAAAGUGAAUGGGUU ..........((.(((((....-------.-.......))))).))(((((((((((.-...((((..(((..-..)))...-----((...)))))).))))))))))).. ( -33.00) >consensus _______________CCACUAU_________UUU_UUUUUUUCGGCCCCAUUCACUUG_GAGGCCUAAGGCUA_AAUCCGCG_____CAACUGGAGGCAAAGUGAAUGGGUU ..............................................(((((((((((.....((((..((....................))..)))).))))))))))).. (-20.37 = -20.73 + 0.36)

| Location | 11,818,024 – 11,818,117 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.94 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.66 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
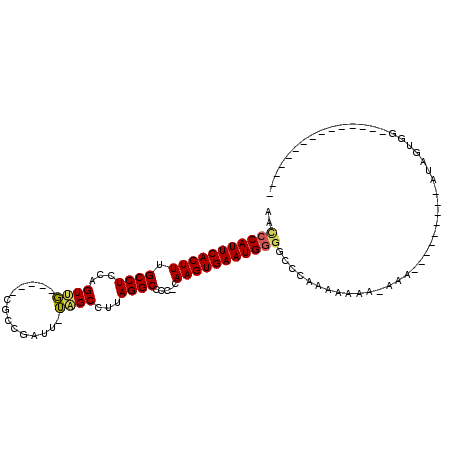
>2R_DroMel_CAF1 11818024 93 - 20766785 AAACCAUUCACUUUGCCUCCAGUUG-----CGCCGAUU-UAGCCUUAGGCCUC-CAAGUGAAUGGGACCGAAAAAAA-GAAAAAACAAACAUAGAGGAGAA----------- ...((((((((((.((((...((((-----........-))))...))))...-.))))))))))............-.......................----------- ( -19.70) >DroPse_CAF1 7462 84 - 1 -UUCCAUUCACUUUGCCUCCAGUCUCCAGUCGCGAUCUUAGGCCUUAGGCCCCACAAGUGAAUGUGGCACACACUCA------------CACUGUGG--------------- -.............((((...((((..((.......)).))))...)))).(((((.((((.((((...)))).)))------------)..)))))--------------- ( -19.70) >DroSec_CAF1 8550 72 - 1 AACCCAUUCACUUUGCCUCCAGUUG-----CGCCGAUU-UAGCUUUAGGCCUC-CAAGUGAAUGGGGCCGAAAAAGA-AA-------------------------------- ..(((((((((((.((((..(((((-----........-)))))..))))...-.)))))))))))...........-..-------------------------------- ( -25.30) >DroSim_CAF1 8468 73 - 1 AACCCAUUCACUUUGCCUCCAGUUG-----CGCCGAUU-UAGCCUUAGGCCUC-CAAGUGAAUGGGGCCGAAAAAGA-AAA------------------------------- ..(((((((((((.((((...((((-----........-))))...))))...-.)))))))))))...........-...------------------------------- ( -23.00) >DroEre_CAF1 8392 85 - 1 AACCCAUUCACUUUGCCUCCAGUUG-----CGCGGCUU-UAGCCUUAGGCCCC-CAAGUGAAUGGGGCCCAAAAGAA-AAA-A-------AUAGUGGAGCC----------- ..(((((((((((.((((...(...-----..)(((..-..)))..))))...-.)))))))))))(((((..(...-...-.-------.)..))).)).----------- ( -28.10) >DroYak_CAF1 8419 97 - 1 AACCCAUUCACUUUGCCUCCAGUGG-----CGCGGCUU-UAGCCUUAGGCCCC-CAAGUGAAUGGGGCCCAACAGAGUAAA-A-------AUAGUGUUGCACAAAAAAAAAA ..(((((((((((.(((......))-----)..(((((-.......)))))..-.)))))))))))...(((((..((...-.-------))..)))))............. ( -30.10) >consensus AACCCAUUCACUUUGCCUCCAGUUG_____CGCCGAUU_UAGCCUUAGGCCCC_CAAGUGAAUGGGGCCCAAAAAAA_AAA_________AUAGUGG_______________ ..(((((((((((.((((...((((..............))))...)))).....))))))))))).............................................. (-18.76 = -18.66 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:45 2006