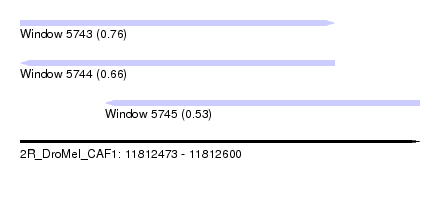
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,812,473 – 11,812,600 |
| Length | 127 |
| Max. P | 0.763244 |

| Location | 11,812,473 – 11,812,573 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11812473 100 + 20766785 GCU--GAGGAGAACGCAUCCAGCUGUGCUGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGAUUGGUGUUGAUGGCCAAUGCGGCGAGC (((--..(((.......))).((((((((((((((((((((((((........)))))))).((((....))))...))))))).))))....))))).))) ( -41.80) >DroVir_CAF1 15462 91 + 1 GGGUGGAGGAC---GCGUCCAGCUGUGCCGCAGCGCCUCGUGUGUCAAUGGCCACACACGAACGCCAUUUGGCGCUGGGUGUUGAUGGCCAGCU-------- .......(((.---...)))(((((.(((((((((((((((((((........)))))))).((((....))))...))))))).)))))))))-------- ( -45.00) >DroPse_CAF1 3113 102 + 1 GAGUGGACGACGACGCUUCCAGCUGUGCUGCAUCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGACUGGUGUUGAUGGCAUAUGCAGAGAAC (((((........)))))...(((((((((((.((((((((((((........)))))))).((((....))))...)))).)).))))))).))....... ( -38.00) >DroMoj_CAF1 18297 94 + 1 GGGUGGACGACGUCGCAUCCAGCUGUGCAGCAGCGCCUCGUGUGUCAAUGGCCACACACGAGCCCCAUUUGGAGCUGGGUGUUGAUGGCCAAUG-------- ...(((....((((((((((((((((((....))))(((((((((........)))))))))..((....))))))))))).))))).)))...-------- ( -41.50) >DroAna_CAF1 3068 100 + 1 GGC--GAGGAGAACGCAUCCAGCUGUGUUGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGAAUGGUGUUGAUGGCCAAUGCAGCGAUC ...--..(((.......))).((((((((((((((((((((((((........)))))))).((((....))))...))))))).....))))))))).... ( -41.30) >DroPer_CAF1 3197 102 + 1 GAGUGGACGACGACGCUUCCAGCUGUGCUGCAUCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGACUGGUGUUGAUGGCAUAUGCAGAGAAC (((((........)))))...(((((((((((.((((((((((((........)))))))).((((....))))...)))).)).))))))).))....... ( -38.00) >consensus GGGUGGACGACGACGCAUCCAGCUGUGCUGCAGCGCCUCGUGUGUCACUUGCCACACACGAACGCCAUUUGGCGAUUGGUGUUGAUGGCCAAUGCAG_GA_C ..............((.....))...(((((((((((((((((((........)))))))).((((....))))...))))))).))))............. (-29.40 = -29.82 + 0.42)

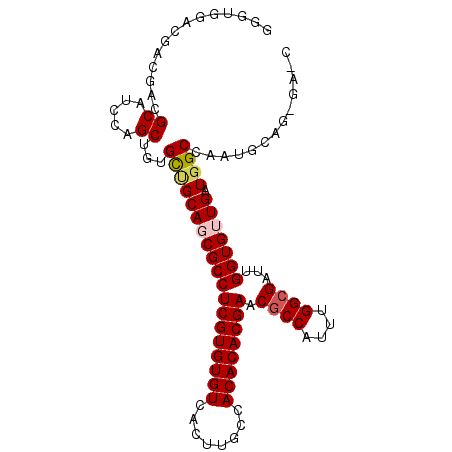
| Location | 11,812,473 – 11,812,573 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -27.31 |
| Energy contribution | -28.98 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11812473 100 - 20766785 GCUCGCCGCAUUGGCCAUCAACACCAAUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCAGCACAGCUGGAUGCGUUCUCCUC--AGC ......((((((.(((............((((....)))).((((((((.(....).)))))))))))((((.....))))..))))))........--... ( -35.90) >DroVir_CAF1 15462 91 - 1 --------AGCUGGCCAUCAACACCCAGCGCCAAAUGGCGUUCGUGUGUGGCCAUUGACACACGAGGCGCUGCGGCACAGCUGGACGC---GUCCUCCACCC --------...(((((((..((((..((((((....)))))).))))))))))).........((((((((.((((...)))).).))---).))))..... ( -38.40) >DroPse_CAF1 3113 102 - 1 GUUCUCUGCAUAUGCCAUCAACACCAGUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGAUGCAGCACAGCUGGAAGCGUCGUCGUCCACUC (((..((((((.((((............((((....)))).((((((((.(....).))))))))))))))))))...)))(((..(((....))))))... ( -36.40) >DroMoj_CAF1 18297 94 - 1 --------CAUUGGCCAUCAACACCCAGCUCCAAAUGGGGCUCGUGUGUGGCCAUUGACACACGAGGCGCUGCUGCACAGCUGGAUGCGACGUCGUCCACCC --------((.(((((((..((((..((((((....)))))).))))))))))).))....((((.((((.(((....))).....))).).))))...... ( -37.40) >DroAna_CAF1 3068 100 - 1 GAUCGCUGCAUUGGCCAUCAACACCAUUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCAACACAGCUGGAUGCGUUCUCCUC--GCC ((..((.(((((.(((............((((....)))).((((((((.(....).)))))))))))((((.....))))..)))))))..))...--... ( -34.40) >DroPer_CAF1 3197 102 - 1 GUUCUCUGCAUAUGCCAUCAACACCAGUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGAUGCAGCACAGCUGGAAGCGUCGUCGUCCACUC (((..((((((.((((............((((....)))).((((((((.(....).))))))))))))))))))...)))(((..(((....))))))... ( -36.40) >consensus G_UC_CUGCAUUGGCCAUCAACACCAAUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGCAGCACAGCUGGAUGCGUCGUCCUCCACCC ........((((((((((..((((....((((....))))...))))))))))))))....((((.(((.(.((((...)))).)))).))))......... (-27.31 = -28.98 + 1.67)



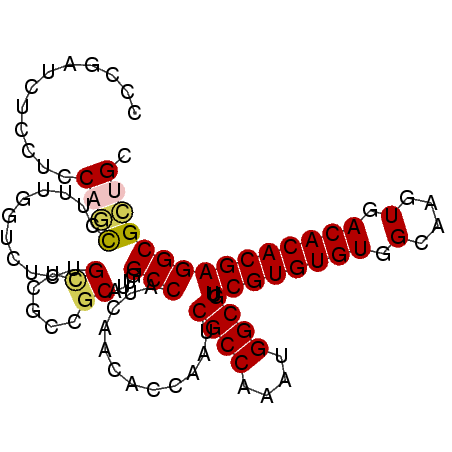
| Location | 11,812,500 – 11,812,600 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11812500 100 - 20766785 CCCGAUCUCCUCCAGUCUUUGGUCUCUGCUCGCCGCAUUGGCCAUCAACACCAAUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGC ............((((...(((((..(((.....)))..)))))......((...((((....)))).((((((((.(....).)))))))))).)))). ( -32.70) >DroPse_CAF1 3142 100 - 1 CUGAAUUGCCUGCUUCUCCUGUUCUCAGUUCUCUGCAUAUGCCAUCAACACCAGUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGAUGC ..((((((...((.......))...))))))...((((.((((............((((....)))).((((((((.(....).)))))))))))))))) ( -33.10) >DroSim_CAF1 3154 100 - 1 CCCGAUCUCCUCCAGUAUUUGGUCUCUGCUCGCCGCAUUGGCCAUCAACACCAAUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGC ............((((...(((((..(((.....)))..)))))......((...((((....)))).((((((((.(....).)))))))))).)))). ( -33.30) >DroEre_CAF1 3170 100 - 1 CCCGAUCUCCUCCAGCCUUUGGACUCUGCUCACCGCAUUGGCCAUCAACACCAAUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGC ............((((.......(..(((.....)))..)(((............((((....)))).((((((((.(....).))))))))))))))). ( -31.10) >DroAna_CAF1 3095 86 - 1 C-------CCCCCAAC-------CUCGGAUCGCUGCAUUGGCCAUCAACACCAUUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGC .-------.......(-------((((..(((((......(((((..((((....((((....))))...))))))))).)))))....)))))...... ( -29.90) >DroPer_CAF1 3226 100 - 1 CUGAAUUGCCUGCUUCUCCUGUUCUCAGUUCUCUGCAUAUGCCAUCAACACCAGUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGAUGC ..((((((...((.......))...))))))...((((.((((............((((....)))).((((((((.(....).)))))))))))))))) ( -33.10) >consensus CCCGAUCUCCUCCAGCCUUUGGUCUCUGCUCGCCGCAUUGGCCAUCAACACCAAUCGCCAAAUGGCGUUCGUGUGUGGCAAGUGACACACGAGGCGCUGC ............((((...........((.....))....(((............((((....)))).((((((((.(....).))))))))))))))). (-25.34 = -25.28 + -0.05)

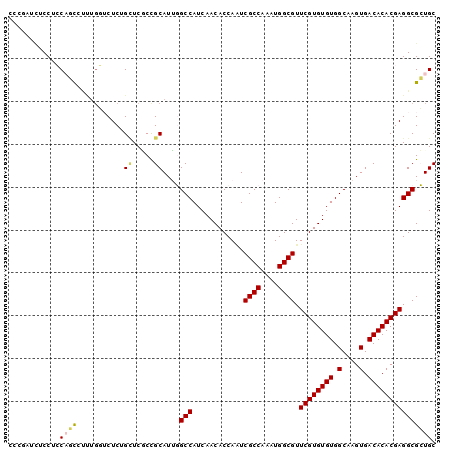
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:43 2006