| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,769,224 – 11,769,354 |
| Length | 130 |
| Max. P | 0.870957 |

| Location | 11,769,224 – 11,769,332 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11769224 108 + 20766785 -----------CGCUGAACUAAAUGAACUCAUCUGCUGAUAAGUUUAGCUGGCCACCGUUUAGCAA-CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUU -----------.(((((......((((((.(((....))).))))))(((((.......)))))..-..........)))))....(((((..(((((......)))))....))))).. ( -27.90) >DroSec_CAF1 13963 109 + 1 -----------ACCUGGUCCAAAUGAACUCAUCUGCUGAAAAGUUUAGCUGGCCACCGUUUAGCAAUCUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUU -----------...(((.(((..((((((..((....))..))))))..))))))..(((..(((........)))...)))....(((((..(((((......)))))....))))).. ( -24.40) >DroSim_CAF1 13290 109 + 1 -----------CCCUGCUCUAAAUGAACUCAUCUGCUGAUAAGUCUAGUUGGCCACCGUUUAGCAAACUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUU -----------.......................(((((((.....((((.((.........)).))))......)))))))....(((((..(((((......)))))....))))).. ( -23.20) >DroEre_CAF1 13599 118 + 1 AGGUGCAGUCACCCUUGACUAAAUGAACUCAUCUGCUGAUAAGU-AAGGUAGUUAUAGUUUAGCAA-CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCACU .(((((((......(((.((((((.((((.(((((((....)))-.))))))))...)))))))))-.....))))))).......(((((..(((((......)))))....))))).. ( -32.10) >DroYak_CAF1 14749 104 + 1 AGGUGCAGACACCCUUGACUAAAUGGACUCAUCUGCUGAUAA---------------GUUUAGCAA-CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACACAACUGCCAUCGCUGGCAUU .((((....))))..((...((((((........((((((((---------------(((....))-))......)))))))......))))))..))......(((((....))))).. ( -22.84) >consensus ___________CCCUGGACUAAAUGAACUCAUCUGCUGAUAAGU_UAGCUGGCCACCGUUUAGCAA_CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUU ..................................(((((((..........((.........))...........)))))))....(((((..(((((......)))))....))))).. (-15.74 = -16.62 + 0.88)

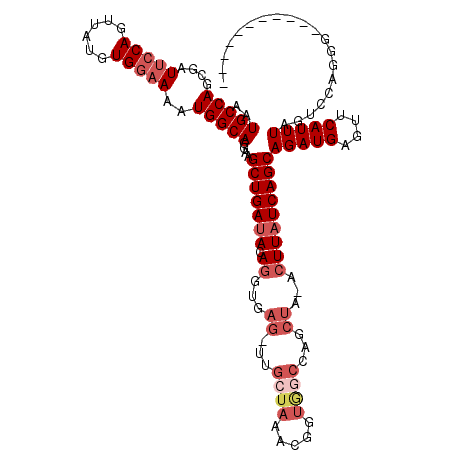
| Location | 11,769,224 – 11,769,332 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -19.96 |
| Energy contribution | -22.04 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11769224 108 - 20766785 AAUGCCAGCGAUUCCAGUUAUGUGGAAAAUGGCAGGAAGCUGAUACAGGUGAG-UUGCUAAACGGUGGCCAGCUAAACUUAUCAGCAGAUGAGUUCAUUUAGUUCAGCG----------- ..(((((....(((((......)))))..)))))....(((((.((((.((.(-(..((....))..)))).)).((((((((....))))))))......))))))).----------- ( -35.40) >DroSec_CAF1 13963 109 - 1 AAUGCCAGCGAUUCCAGUUAUGUGGAAAAUGGCAGGAAGCUGAUACAGGUGAGAUUGCUAAACGGUGGCCAGCUAAACUUUUCAGCAGAUGAGUUCAUUUGGACCAGGU----------- ..(((((....(((((......)))))..)))))((..(((((...((...((...((((.....))))...))...))..)))))(((((....)))))...))....----------- ( -28.90) >DroSim_CAF1 13290 109 - 1 AAUGCCAGCGAUUCCAGUUAUGUGGAAAAUGGCAGGAAGCUGAUACAGGUGAGUUUGCUAAACGGUGGCCAACUAGACUUAUCAGCAGAUGAGUUCAUUUAGAGCAGGG----------- ..(((((....(((((......)))))..)))))....(((((((.((...((((.((((.....)))).))))...)))))))))......((((.....))))....----------- ( -33.30) >DroEre_CAF1 13599 118 - 1 AGUGCCAGCGAUUCCAGUUAUGUGGAAAAUGGCAGGAAGCUGAUACAGGUGAG-UUGCUAAACUAUAACUACCUU-ACUUAUCAGCAGAUGAGUUCAUUUAGUCAAGGGUGACUGCACCU .(((.((((...(((.(((((.......))))).))).)))).)))(((((((-(..((........((((....-(((((((....))))))).....))))....))..))).))))) ( -35.90) >DroYak_CAF1 14749 104 - 1 AAUGCCAGCGAUGGCAGUUGUGUGGAAAAUGGCAGGAAGCUGAUACAGGUGAG-UUGCUAAAC---------------UUAUCAGCAGAUGAGUCCAUUUAGUCAAGGGUGUCUGCACCU ..(((((....)))))((((.(((((...((((((...(((......)))...-))))))..(---------------(((((....))))))))))).))).)..(((((....))))) ( -31.10) >consensus AAUGCCAGCGAUUCCAGUUAUGUGGAAAAUGGCAGGAAGCUGAUACAGGUGAG_UUGCUAAACGGUGGCCAGCUA_ACUUAUCAGCAGAUGAGUUCAUUUAGUCCAGGG___________ ..(((((....(((((......)))))..)))))....(((((((.((...((...((((.....))))...))...)))))))))(((((....))))).................... (-19.96 = -22.04 + 2.08)

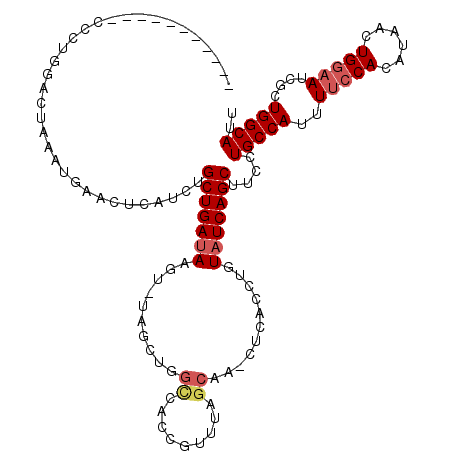
| Location | 11,769,253 – 11,769,354 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.45 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11769253 101 + 20766785 AAGUUUAGCUGGCCACCGUUUAGCAA-CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUUUGGCUGAUAGU---------GCUGUUUUUGC .......(((((.......)))))((-(.(((...((((((((...(((((..(((((......)))))....)))))...))))))))))---------)..)))..... ( -27.80) >DroSec_CAF1 13992 102 + 1 AAGUUUAGCUGGCCACCGUUUAGCAAUCUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUUUGGCUGGUAGU---------GCUGUUUUUGC ......................((((...((.(..((((((((...(((((..(((((......)))))....)))))...))))))))..---------).))...)))) ( -26.70) >DroSim_CAF1 13319 102 + 1 AAGUCUAGUUGGCCACCGUUUAGCAAACUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUUUGGCUGGUAGU---------GCUGUUUUUGC ..(((.....))).........(((((..((.(..((((((((...(((((..(((((......)))))....)))))...))))))))..---------).))..))))) ( -29.50) >DroEre_CAF1 13639 100 + 1 AAGU-AAGGUAGUUAUAGUUUAGCAA-CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCACUUGGCUGGUAGU---------GCUGUUUUUGC .(((-(((((((((..........))-)).)))).((((((((...(((((..(((((......)))))....)))))...)))))))).)---------)))........ ( -29.00) >DroYak_CAF1 14789 95 + 1 AA---------------GUUUAGCAA-CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACACAACUGCCAUCGCUGGCAUUUGGCUGGUAGUGAUGCUUCUGCUGUUUUUGC .(---------------((..((((.-.((((...((((((((...(((((.......((....)).......)))))...))))))))))))))))...)))........ ( -24.84) >consensus AAGU_UAGCUGGCCACCGUUUAGCAA_CUCACCUGUAUCAGCUUCCUGCCAUUUUCCACAUAACUGGAAUCGCUGGCAUUUGGCUGGUAGU_________GCUGUUUUUGC ....................((((...........((((((((...(((((..(((((......)))))....)))))...))))))))...........))))....... (-23.21 = -23.45 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:23 2006