| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,758,996 – 11,759,098 |
| Length | 102 |
| Max. P | 0.754603 |

| Location | 11,758,996 – 11,759,098 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -25.73 |
| Energy contribution | -25.15 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
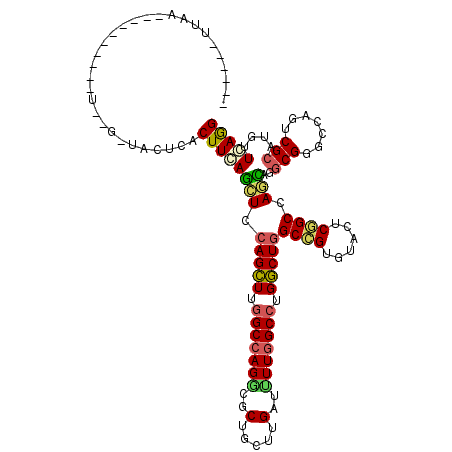
>2R_DroMel_CAF1 11758996 102 + 20766785 ------UCUA----------U-AGAUACUCACUUCAUCUCCAGCUUCGCGAGGCGCUGCUUGAUCUUGGCCUGGCUUGCCGUGUACUCAGCCAGGAGGCGGGCCAGCCGCAUGUUGAGG ------....----------.-.....((((..(((....((((.((....)).))))..)))....(((.(((((((((.((........))...))))))))))))......)))). ( -34.30) >DroVir_CAF1 206 104 + 1 ------UUAAGUCACA---------AACUCACUUGAGCUCCAGCUUGGCCAGGCGCUGUUUGAUUUUGGCCUGGCUGGCCGUGUACUCGGCAAGCAGGCGGGCCAGUCGCAUGUUCAAG ------..........---------......(((((((.((((((.(((((((..(.....)..))))))).))))))..(((.(((.(((..(....)..)))))))))..))))))) ( -39.60) >DroGri_CAF1 5300 102 + 1 ------UUAAGGC--C---------AACUCACUUGAGCUCCAGUUUGGCCAGACGCUGCUUGAUUUUGGCCUGACUGGCCGUAUAUUCGGCCAACAGACGGGCCAGACGCAUGUUGAGG ------....(((--(---------(((((....))).......)))))).((((.(((.....(((((((((..((((((......)))))).....))))))))).))))))).... ( -38.01) >DroWil_CAF1 2118 108 + 1 ACAUUAGGAA----------U-UGUCACUUACUUCAUUUCCAGUUUGGCCAGGCGCUGUUUGAUUUUGGCCUGACUGGCCGUAUACUCGGCCAAGAGGCGUGCCAAACGCAUAUUGAGG ......((((----------.-((..........)).)))).((((((((((((...)))))......((((...((((((......))))))..))))..)))))))........... ( -34.10) >DroMoj_CAF1 4180 112 + 1 ------AUGAGCCGAAUGGCU-GGCUACUCACUUGAGCUCCAGCUUGGCCAGACGCUGCUUGAUUUUGGCCUGGCUGGCCGUGUACUCGGCCAGCAGGCGCGCCAGUCGCAUGUUCAGG ------.(((((....(((((-(((..(((....))).....(((((((((((..(.....)..)))))))..((((((((......))))))))))))..))))))))...))))).. ( -51.50) >DroAna_CAF1 5240 103 + 1 ------UCUA----------UGCGGUACUCACUUCAUCUCCAGCUUGGCCAGUCGCUGCUUGAUCUUGGCCUGGCUGGCGGUGUAUUCCGCCAGAAGGCGCGCCAGUCGCAUGUUCAGG ------..((----------(((((.........((((.((((((.(((((((((.....))))..))))).)))))).)))).....((((....))))......)))))))...... ( -38.80) >consensus ______UUAA__________U__G_UACUCACUUCAGCUCCAGCUUGGCCAGGCGCUGCUUGAUUUUGGCCUGGCUGGCCGUGUACUCGGCCAGCAGGCGGGCCAGUCGCAUGUUCAGG ...............................((((((((.(((((.(((((((..(.....)..))))))).)))))((((......)))).)))..(((.......)))....))))) (-25.73 = -25.15 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:19 2006