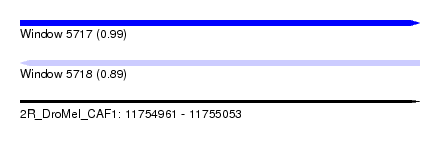
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,754,961 – 11,755,053 |
| Length | 92 |
| Max. P | 0.989100 |

| Location | 11,754,961 – 11,755,053 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -19.91 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
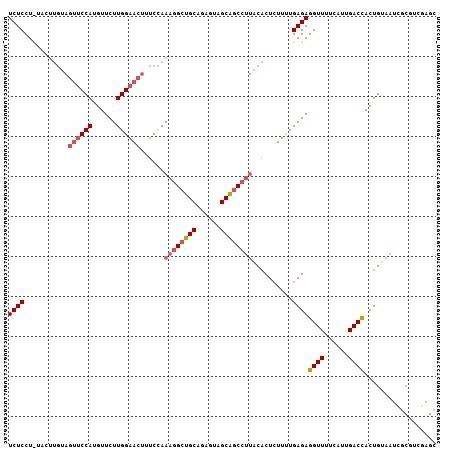
>2R_DroMel_CAF1 11754961 92 + 20766785 UCUCCU------------CCAUGUUCUUGGAAACUGCUAAAGGCUGCGGAGUAGCGUC---UCUCUCUUCUGAGAGGUUUUCAUUGACCACUGAUAUUGCGUCGAGC .(((((------------(((......))))....((........))))))..((.((---((((......))))))......(((((((.......)).))))))) ( -22.90) >DroSec_CAF1 7456 107 + 1 UCUCCUUUACUUGUAGUUCCAUGUUCUUGGAACUUUCCAAAGGCUGCAGAGUAGCAGCCUUACACUCUUUUGAGAGGUUUUCAUUGACUACUGUAAUGANNNNNNNN ...(((.....((((((((((......))))))).....((((((((......)))))))))))(((....))))))...((((((.(....)))))))........ ( -29.00) >DroSim_CAF1 6666 107 + 1 UCUCCUCUACUUGUAGUUCCAUGUUCUUGGAACUUUCCAAAGGCUGCAGAGUAGCAGCCUUACACUGUUUUGAGAGGUUUUCAUUGACCACUGUAAUCGCGUCGAGC .((((((.((.((((((((((......))))))).....((((((((......)))))))))))..))...))).((((......))))..............))). ( -32.10) >consensus UCUCCU_UACUUGUAGUUCCAUGUUCUUGGAACUUUCCAAAGGCUGCAGAGUAGCAGCCUUACACUCUUUUGAGAGGUUUUCAUUGACCACUGUAAUCGCGUCGAGC ((((...........((((((......))))))......((((((((......))))))))..........))))((((......)))).................. (-19.91 = -21.80 + 1.89)

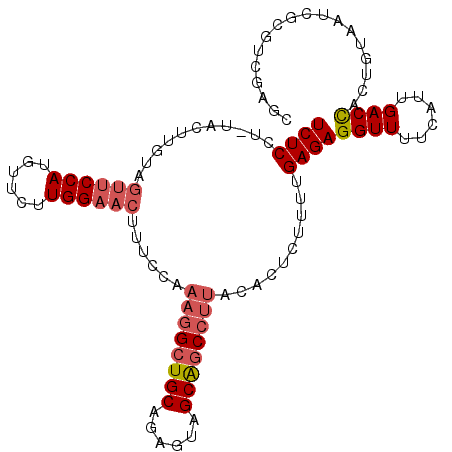
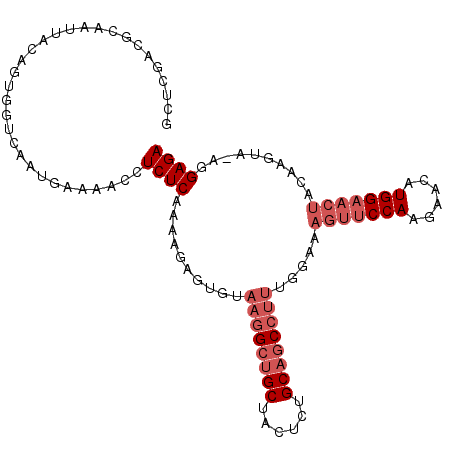
| Location | 11,754,961 – 11,755,053 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -14.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11754961 92 - 20766785 GCUCGACGCAAUAUCAGUGGUCAAUGAAAACCUCUCAGAAGAGAGA---GACGCUACUCCGCAGCCUUUAGCAGUUUCCAAGAACAUGG------------AGGAGA (((((((.((.......))))).........(((((....))))).---)).))..((((((........))....((((......)))------------))))). ( -22.70) >DroSec_CAF1 7456 107 - 1 NNNNNNNNUCAUUACAGUAGUCAAUGAAAACCUCUCAAAAGAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAAAGGAGA ........(((((((....)).))))).....((((.......(((((((((((......)))))))).....(((((((......)))))))))).......)))) ( -29.74) >DroSim_CAF1 6666 107 - 1 GCUCGACGCGAUUACAGUGGUCAAUGAAAACCUCUCAAAACAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUAGAGGAGA ....(((((.......)).)))........((((((..........((((((((......)))))))).....(((((((......)))))))....).)))))... ( -32.50) >consensus GCUCGACGCAAUUACAGUGGUCAAUGAAAACCUCUCAAAAGAGUGUAAGGCUGCUACUCUGCAGCCUUUGGAAAGUUCCAAGAACAUGGAACUACAAGUA_AGGAGA ................................((((..........((((((((......)))))))).....(((((((......)))))))..........)))) (-14.90 = -17.90 + 3.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:16 2006