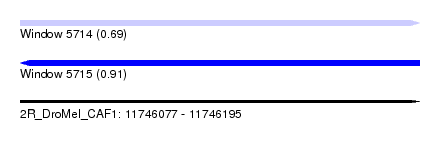
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,746,077 – 11,746,195 |
| Length | 118 |
| Max. P | 0.913816 |

| Location | 11,746,077 – 11,746,195 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -27.88 |
| Energy contribution | -29.00 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11746077 118 + 20766785 AUCAAAAGAAUAUUCCCCUGUUGCACAGACUCAAAAGGAGCACAAGCCACAGUCUCCUUCCAAGGGAGAUGCUCGUAUUGUUCCGGCAAGAUCCAGAACUUAUACACGCACCUUGGAU .............(((.(((.....)))......((((.((...(((....(((((((.....)))))))))).((((.((((.((......)).))))..))))..)).))))))). ( -29.40) >DroSec_CAF1 12730 118 + 1 AUCAAAAGAAUAUUCCCCUGUUGCACAGGCUAAAAAGGAGCACAAGCCCCCGUCUCCUUCCAAGGGAGAUGCUCGUGUUGUUCCAGCAAGAUCCAGAACUUACACCCGCACCUUGGAU .............(((((((.....)))).....((((.((.........((((((((.....))))))))...((((.((((............))))..))))..)).))))))). ( -30.10) >DroSim_CAF1 8954 118 + 1 AUCAAAAGAAUAUUCCCCUGUUGCACAGGCUAAAAAGGAGCACAAGCCCCCGUCUCCUUCCAAGGGAGAUGCUCGUGUUGUUCCGGCGAGAGCCAGAACUUAUACCCGCACCUUGGAU .............(((((((.....)))).....(((((((((.(((....(((((((.....)))))))))).)))))((((.(((....))).))))...........))))))). ( -39.10) >DroEre_CAF1 22583 118 + 1 UUCAAAGGAAUAUUCCCCUGUUGCUCAGGCUAAAAAGGAGCACAAGUCCAACUCUCUUUCCAAAGGAGAUGCUCGUGCUGUUCCGGCAAGAGCCAGAACUUAUACCCGCACCUUGGAU .((.((((........((((.....)))).......(((((((.(((.....((((((.....)))))).))).)))))((((.(((....))).))))......))...)))).)). ( -32.70) >DroYak_CAF1 13132 118 + 1 UCCUAAGGCAUAUUCCCCUGUUGCUCAGCCCAAAAAGGAGCACAAGUCCAACUCUCCUGGCAAAGGCGACGCUCGUGCUAUUCCGGCAAGGGCCAGAACUUAUACCCGCACCUUGGAU ..((((((...........((((((..(((.....(((((....((.....))))))))))...))))))((((.((((.....)))).)))).................)))))).. ( -31.90) >consensus AUCAAAAGAAUAUUCCCCUGUUGCACAGGCUAAAAAGGAGCACAAGCCCCAGUCUCCUUCCAAGGGAGAUGCUCGUGUUGUUCCGGCAAGAGCCAGAACUUAUACCCGCACCUUGGAU .............(((((((.....)))).....(((((((((.(((....(((((((.....)))))))))).)))))((((.(((....))).))))...........))))))). (-27.88 = -29.00 + 1.12)

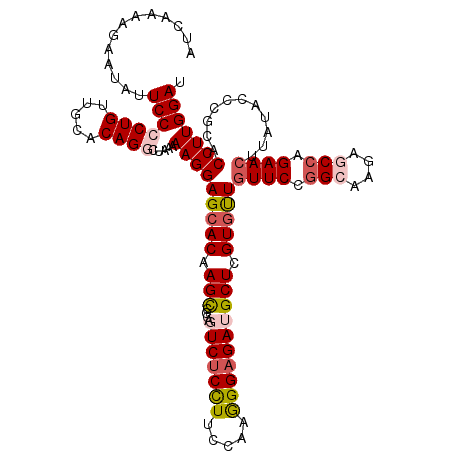
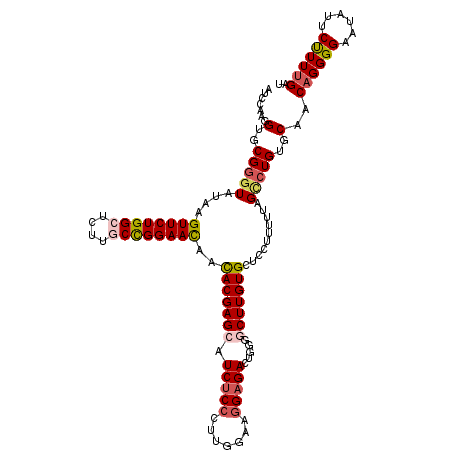
| Location | 11,746,077 – 11,746,195 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -32.52 |
| Energy contribution | -33.48 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11746077 118 - 20766785 AUCCAAGGUGCGUGUAUAAGUUCUGGAUCUUGCCGGAACAAUACGAGCAUCUCCCUUGGAAGGAGACUGUGGCUUGUGCUCCUUUUGAGUCUGUGCAACAGGGGAAUAUUCUUUUGAU .(((.((((((.(((((..(((((((......))))))).))))).))))))..((..(((((((((........)).)))))))..))((((.....)))))))............. ( -41.00) >DroSec_CAF1 12730 118 - 1 AUCCAAGGUGCGGGUGUAAGUUCUGGAUCUUGCUGGAACAACACGAGCAUCUCCCUUGGAAGGAGACGGGGGCUUGUGCUCCUUUUUAGCCUGUGCAACAGGGGAAUAUUCUUUUGAU .((((((((((..((((..((((..(......)..)))).))))..)))....)))))))((((((.((((((....))))))......((((.....)))).......))))))... ( -42.00) >DroSim_CAF1 8954 118 - 1 AUCCAAGGUGCGGGUAUAAGUUCUGGCUCUCGCCGGAACAACACGAGCAUCUCCCUUGGAAGGAGACGGGGGCUUGUGCUCCUUUUUAGCCUGUGCAACAGGGGAAUAUUCUUUUGAU .......(..(((((....((((((((....)))))))).....((((((..((((((........))))))...)))))).......)))))..)..(((((((....))))))).. ( -46.60) >DroEre_CAF1 22583 118 - 1 AUCCAAGGUGCGGGUAUAAGUUCUGGCUCUUGCCGGAACAGCACGAGCAUCUCCUUUGGAAAGAGAGUUGGACUUGUGCUCCUUUUUAGCCUGAGCAACAGGGGAAUAUUCCUUUGAA .......((.(((((....((((((((....))))))))(((((((((..(((.(((....))))))...).))))))))........))))).))..(((((((....))))))).. ( -45.20) >DroYak_CAF1 13132 118 - 1 AUCCAAGGUGCGGGUAUAAGUUCUGGCCCUUGCCGGAAUAGCACGAGCGUCGCCUUUGCCAGGAGAGUUGGACUUGUGCUCCUUUUUGGGCUGAGCAACAGGGGAAUAUGCCUUAGGA .((((((((((..((....((((((((....))))))))...))..)).((.((((((((((..(((..((((....).)))..)))...))).)))).))).))....))))).))) ( -39.30) >consensus AUCCAAGGUGCGGGUAUAAGUUCUGGCUCUUGCCGGAACAACACGAGCAUCUCCCUUGGAAGGAGACUGGGGCUUGUGCUCCUUUUUAGCCUGUGCAACAGGGGAAUAUUCUUUUGAU .......(..(((((....((((((((....))))))))..(((((((.(((((.......))))).....)))))))..........)))))..)..((((((......)))))).. (-32.52 = -33.48 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:13 2006