
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
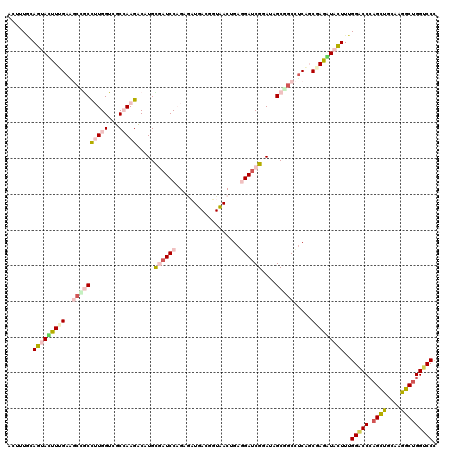
| Location | 11,742,881 – 11,743,001 |
| Length | 120 |
| Max. P | 0.851207 |

| Location | 11,742,881 – 11,743,001 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -21.69 |
| Energy contribution | -23.42 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
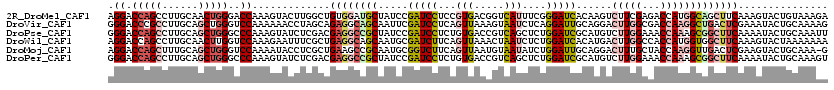
>2R_DroMel_CAF1 11742881 120 + 20766785 UCUUUACAGUACUUUGAAGCUGCCAUGGUCUCGAAGACUUGUGAUCCCGAAAUGACCGUCACGGAGGAUCGGAUAGCAUCCACAGCCAAGUACUUUGGUCCCAGUUGCAAGGCUGGUCCU ...(((((((.((((((..((.....))..)))))))).)))))..(((..((....))..)))(((((.((((...)))).(((((..(((..(((....))).)))..)))))))))) ( -34.10) >DroVir_CAF1 12518 120 + 1 CUUUUGCAGUAUUUCGAGUCAGCCUUGGUCGCCAAGUCCUGCAAUCCUGAGAUUACUUUAACUGAGGAUCGAAUUGCUGCCUCUGCUAGGUUUUUUGGACCCAGCUGCAAGGCGGGUCCC .....((((((.((((((.(((.(((((...)))))..))))..((((.((..........)).))))))))).))))))................((((((.(((....))))))))). ( -41.20) >DroPse_CAF1 5462 120 + 1 AAUUUGCAGUAUUUUGAAGCCGCUUUGGUUUCCAAGACAUGCGAUCCAGAGCUGACGGUCACAGAGGAUCGGAUAGCGGCCUCGUCGAGAUACUUUGGGCCCAGCUGCAAGGCUGGUCCC .......((((((((((.((((((((((...))))...((.((((((....(((.......))).)))))).))))))))....))))))))))..(((.((((((....)))))).))) ( -47.40) >DroWil_CAF1 34094 120 + 1 UUUUUUUAGUACUUUGAAGCCACCAUGGUGGCCAAGUCAUGUGAUCCAGAGAUUAGUUUAACUGAAGAUCGCAUUGCUGCCUCAGCGAAAUUCUUUGGACCAAGUUGCAAGGCUGGUCCU ..............(((.(((((....)))))....)))((((((((((..(......)..)))..)))))))((((((...))))))........((((((.(((....))))))))). ( -34.70) >DroMoj_CAF1 23218 119 + 1 C-UUUGCAGUACUUCGAGUCAACCUUGGUAGCAAAGUCCUGCAAUCCAGAUAUUACAUUAACUGAAGACCGCAUUGCGGCUUCAGCGAGGUAUUUUGGACCCAGCUGCAAAGCUGGUCCU .-.....(((((((((........((((..(((......)))...))))............((((((.((((...)))))))))))))))))))..(((((.((((....))))))))). ( -42.60) >DroPer_CAF1 5679 120 + 1 ACUUUGCAGUAUUUUGAAGCCGCUUUGGUUUCCAAGACAUGCGAUCCAGAGCUGACGGUCACAGAGGAUCGGAUAGCGGCCUCGUCGAGAUACUUUGGGCCCAGCUGCAAGGCUGGUCCC .......((((((((((.((((((((((...))))...((.((((((....(((.......))).)))))).))))))))....))))))))))..(((.((((((....)))))).))) ( -47.40) >consensus ACUUUGCAGUACUUUGAAGCCGCCUUGGUCGCCAAGACAUGCGAUCCAGAGAUGACGGUAACUGAGGAUCGGAUAGCGGCCUCAGCGAGAUACUUUGGACCCAGCUGCAAGGCUGGUCCC .......(((((((((..((((((((((...))))).....((((((..................))))))....))))).....)))))))))..(((((.((((....))))))))). (-21.69 = -23.42 + 1.73)


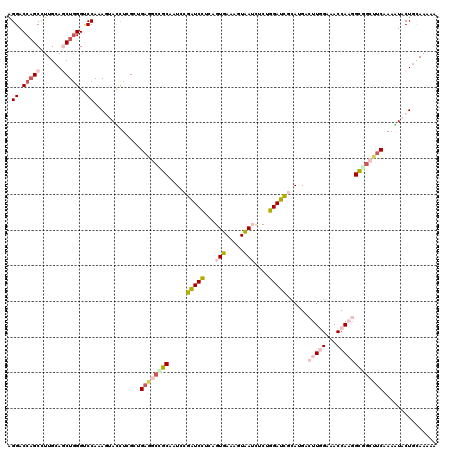
| Location | 11,742,881 – 11,743,001 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.75 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11742881 120 - 20766785 AGGACCAGCCUUGCAACUGGGACCAAAGUACUUGGCUGUGGAUGCUAUCCGAUCCUCCGUGACGGUCAUUUCGGGAUCACAAGUCUUCGAGACCAUGGCAGCUUCAAAGUACUGUAAAGA .((.((((........))))..))..(((((((((((((((((...))))((((((..(((.....)))...))))))....(((.....)))....))))))...)))))))....... ( -35.70) >DroVir_CAF1 12518 120 - 1 GGGACCCGCCUUGCAGCUGGGUCCAAAAAACCUAGCAGAGGCAGCAAUUCGAUCCUCAGUUAAAGUAAUCUCAGGAUUGCAGGACUUGGCGACCAAGGCUGACUCGAAAUACUGCAAAAG .......(((((...(((((((.......))))))).))))).(((.(((((...(((((....((((((....))))))....(((((...)))))))))).)))))....)))..... ( -39.90) >DroPse_CAF1 5462 120 - 1 GGGACCAGCCUUGCAGCUGGGCCCAAAGUAUCUCGACGAGGCCGCUAUCCGAUCCUCUGUGACCGUCAGCUCUGGAUCGCAUGUCUUGGAAACCAAAGCGGCUUCAAAAUACUGCAAAUU (((.(((((......))))).)))..(((((......(((((((((...((((((.(((.......)))....))))))......((((...)))))))))))))...)))))....... ( -43.10) >DroWil_CAF1 34094 120 - 1 AGGACCAGCCUUGCAACUUGGUCCAAAGAAUUUCGCUGAGGCAGCAAUGCGAUCUUCAGUUAAACUAAUCUCUGGAUCACAUGACUUGGCCACCAUGGUGGCUUCAAAGUACUAAAAAAA .(((((((.........)))))))..........((((...)))).(((.(((((..((..........))..))))).))).((((((((((....))))))...)))).......... ( -32.80) >DroMoj_CAF1 23218 119 - 1 AGGACCAGCUUUGCAGCUGGGUCCAAAAUACCUCGCUGAAGCCGCAAUGCGGUCUUCAGUUAAUGUAAUAUCUGGAUUGCAGGACUUUGCUACCAAGGUUGACUCGAAGUACUGCAAA-G .(((((((((....)))).)))))....(((...(((((((((((...)))).)))))))....))).........((((((.((((((..((....)).....)))))).)))))).-. ( -43.10) >DroPer_CAF1 5679 120 - 1 GGGACCAGCCUUGCAGCUGGGCCCAAAGUAUCUCGACGAGGCCGCUAUCCGAUCCUCUGUGACCGUCAGCUCUGGAUCGCAUGUCUUGGAAACCAAAGCGGCUUCAAAAUACUGCAAAGU (((.(((((......))))).)))..(((((......(((((((((...((((((.(((.......)))....))))))......((((...)))))))))))))...)))))....... ( -43.10) >consensus AGGACCAGCCUUGCAGCUGGGUCCAAAGUACCUCGCUGAGGCCGCAAUCCGAUCCUCAGUGAAAGUAAUCUCUGGAUCGCAUGACUUGGAAACCAAGGCGGCUUCAAAAUACUGCAAAAA .((.(((((......)))))..)).............((((((((.....(((((...(((.....)))....)))))......(((((...)))))))))))))............... (-20.27 = -21.75 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:10 2006