| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,306,898 – 2,307,023 |
| Length | 125 |
| Max. P | 0.993816 |

| Location | 2,306,898 – 2,306,992 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2306898 94 + 20766785 CUACUAGGCCCUUUCUCUUUGGGCGGGGCAAACAAAAGGCAGCGGUCUGCAAAUGUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCU .......((((.........)))).(((((((((....((((....))))...))))))))-).(((..(((((((.....)))))))..))).. ( -37.80) >DroSec_CAF1 78079 94 + 1 CUACUUGCCCCUUUCUCUUUGGGCGGGGCAAACAAAAGGCAGCGGGCUGCAAAUGUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCU .....(((((..........)))))(((((((((....((((....))))...))))))))-).(((..(((((((.....)))))))..))).. ( -38.60) >DroSim_CAF1 74614 94 + 1 CUACUUGCCCCUUUUUCUUUGGGCGGGGCAAACAAAAGGCAGCGGGCUGCAAAUGUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCU .....(((((..........)))))(((((((((....((((....))))...))))))))-).(((..(((((((.....)))))))..))).. ( -38.60) >DroEre_CAF1 72283 94 + 1 CUACUGGGCCCUUUCUCUUUGAGCGAGGCAAACAAAAGGCGGCGAGUUGCAAAUAUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUGAGCCCU ...((((((((((.(((...))).))))..........((((....))))........)))-)))((.((((((((.....)))))))).))... ( -30.80) >DroYak_CAF1 68221 83 + 1 CUG-----------CUCUUUGGGCGGGGCAAACAAAAGGCAGCGAAUUGCAAAUAUUUGGC-CAGGCACAAGGCAAACUGUUAGCUUAAAGCCCU ...-----------......((((.((((.((((....((((....)))).........((-(........)))....)))).))))...)))). ( -22.50) >DroAna_CAF1 93603 91 + 1 --ACUUGGUCCU--GUGUUUGAGCAGGACAAACACAGUGCAGAAAAUUGCAAAUAUUUGCCCCGGGCACAGACCAAACAGUUAGCUUUAAGCCUU --..((((((((--(((((((.......)))))))))(((((....)))))......((((...))))..)))))).......((.....))... ( -24.70) >consensus CUACUUGGCCCUUUCUCUUUGGGCGGGGCAAACAAAAGGCAGCGAGCUGCAAAUAUUUGCC_CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCU ....................((((.((((((((.....((((....)))).......((((...))))...........))))))))...)))). (-19.72 = -20.05 + 0.33)

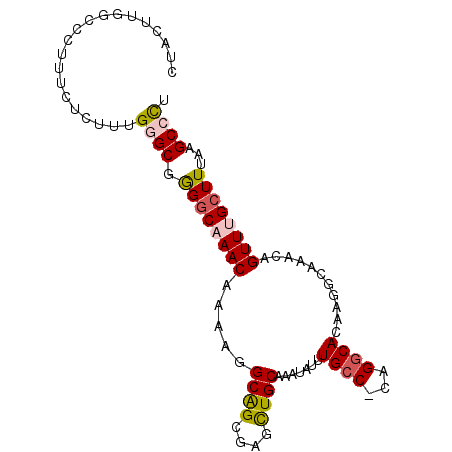
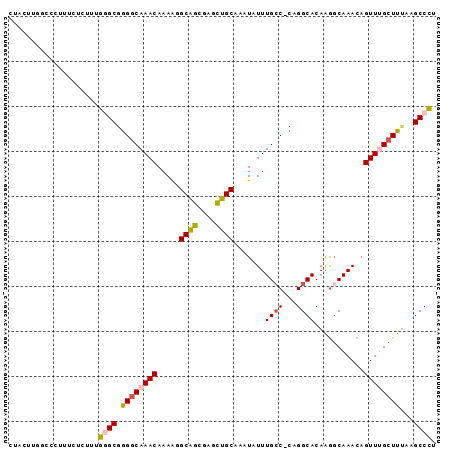
| Location | 2,306,898 – 2,306,992 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2306898 94 - 20766785 AGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAACAUUUGCAGACCGCUGCCUUUUGUUUGCCCCGCCCAAAGAGAAAGGGCCUAGUAG .((((.(((.((((.....)))).))).))))(-((((((((...((((....))))....))))))))).((((.........))))....... ( -35.90) >DroSec_CAF1 78079 94 - 1 AGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAACAUUUGCAGCCCGCUGCCUUUUGUUUGCCCCGCCCAAAGAGAAAGGGGCAAGUAG .(((((....(((((.(((((((((.......)-))))))))))))))))))...........((((((((.............))))))))... ( -39.02) >DroSim_CAF1 74614 94 - 1 AGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAACAUUUGCAGCCCGCUGCCUUUUGUUUGCCCCGCCCAAAGAAAAAGGGGCAAGUAG .(((((....(((((.(((((((((.......)-))))))))))))))))))...........((((((((.............))))))))... ( -39.02) >DroEre_CAF1 72283 94 - 1 AGGGCUCAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAAUAUUUGCAACUCGCCGCCUUUUGUUUGCCUCGCUCAAAGAGAAAGGGCCCAGUAG .((((((...(((((((((((((((.......)-))))))))...((.....))........))))))(((.......)))...))))))..... ( -32.40) >DroYak_CAF1 68221 83 - 1 AGGGCUUUAAGCUAACAGUUUGCCUUGUGCCUG-GCCAAAUAUUUGCAAUUCGCUGCCUUUUGUUUGCCCCGCCCAAAGAG-----------CAG .((((.....((.(((((((((((........)-).)))))....(((......)))....)))).))...))))......-----------... ( -18.50) >DroAna_CAF1 93603 91 - 1 AAGGCUUAAAGCUAACUGUUUGGUCUGUGCCCGGGGCAAAUAUUUGCAAUUUUCUGCACUGUGUUUGUCCUGCUCAAACAC--AGGACCAAGU-- ..(((.....))).....(((((((((((..((((((((((((.((((......))))..))))))))))))......)))--).))))))).-- ( -32.70) >consensus AGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG_GGCAAACAUUUGCAACCCGCUGCCUUUUGUUUGCCCCGCCCAAAGAGAAAGGGCCAAGUAG .((((.....((((((((((((((..........))))))))...(((......))).....))))))...)))).................... (-19.20 = -19.62 + 0.42)
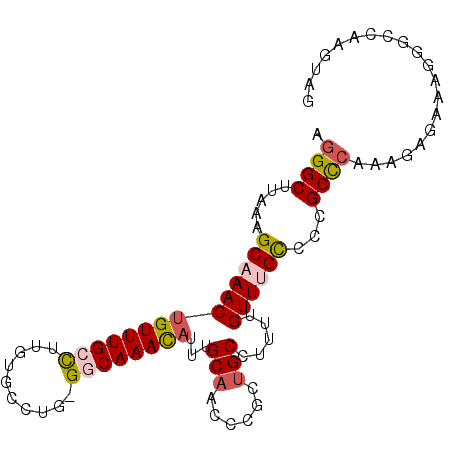
| Location | 2,306,931 – 2,307,023 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.15 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -12.21 |
| Energy contribution | -12.24 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2306931 92 + 20766785 AAAAGGC-----AGCGGUCUGCAAAUGUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCUUGCCACUUGACCUUUCU---UUU-UUCCCGCUGCC----- ....(((-----(((((...(((((((((((((-.........)))))))).))))).(((((.........))))).......---...-...))))))))----- ( -32.00) >DroPse_CAF1 73142 107 + 1 AGGAGGGAGGGGAGUGAACUGCAAAUAUUUGCCCCAGGCACAGGGCAAACAAUCCGCUCAAAGCCUUUGCCACUUGACCUUCUCCCCUUUUUUCCUGCUCCCCAGAA .(((((((((((((......((((....))))....((((.(((((................)))))))))..........)))))))))))))(((.....))).. ( -33.59) >DroSim_CAF1 74647 93 + 1 AAAAGGC-----AGCGGGCUGCAAAUGUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCUUGCCACUUGACCUUCCU---UUUUUUGCCACUGCC----- ....(((-----((.((((((((((((((((((-.........)))))))).)))))....)))))))))).............---...............----- ( -31.70) >DroEre_CAF1 72316 92 + 1 AAAAGGC-----GGCGAGUUGCAAAUAUUUGCC-CAGGCACAAGGCAAACAGUUUGCUUUGAGCCCUUGCCGCCUGACCUUUCU---UUU-UUGCCGCUGCC----- ...((((-----((((((..((((....)))).-..(((.((((((((.....)))))))).))))))))))))).........---...-...........----- ( -35.50) >DroYak_CAF1 68243 91 + 1 AAAAGGC-----AGCGAAUUGCAAAUAUUUGGC-CAGGCACAAGGCAAACUGUUAGCUUAAAGCCCUUGCCAGUUGACCUUUUC---UUU-UU-UUGCUGCC----- ....(((-----((((((..........(((((-..(((...((((.........))))...)))...)))))..((.....))---...-.)-))))))))----- ( -24.60) >DroAna_CAF1 93632 94 + 1 ACAGUGC-----AGAAAAUUGCAAAUAUUUGCCCCGGGCACAGACCAAACAGUUAGCUUUAAGCCUUUGCCACUUGACCUUCCC---UUUUUCGACGCUGCU----- .(((((.-----.(((((..((((....))))....((((..(((......))).((.....))...)))).............---.)))))..)))))..----- ( -18.30) >consensus AAAAGGC_____AGCGAACUGCAAAUAUUUGCC_CAGGCACAAGGCAAACAGUUUGCUUUAAGCCCUUGCCACUUGACCUUCCC___UUU_UUGCCGCUGCC_____ .(((((.......(((((..((((....))))....(((..(((((.........)))))..)))))))).......)))))......................... (-12.21 = -12.24 + 0.03)

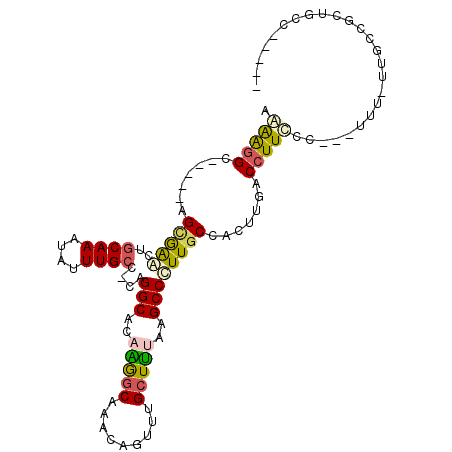
| Location | 2,306,931 – 2,307,023 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.15 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2306931 92 - 20766785 -----GGCAGCGGGAA-AAA---AGAAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAACAUUUGCAGACCGCU-----GCCUUUU -----((((((((...-..(---((....(((....))).....)))...(((((.(((((((((.......)-)))))))))))))...)))))-----))).... ( -39.80) >DroPse_CAF1 73142 107 - 1 UUCUGGGGAGCAGGAAAAAAGGGGAGAAGGUCAAGUGGCAAAGGCUUUGAGCGGAUUGUUUGCCCUGUGCCUGGGGCAAAUAUUUGCAGUUCACUCCCCUCCCUCCU .....(((((..(((.....((((((....((((..(((....)))))))((((((.(((((((((......)))))))))))))))......)))))))))))))) ( -42.50) >DroSim_CAF1 74647 93 - 1 -----GGCAGUGGCAAAAAA---AGGAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAACAUUUGCAGCCCGCU-----GCCUUUU -----((((((.........---......(((....)))..(((((....(((((.(((((((((.......)-)))))))))))))))))))))-----))).... ( -38.80) >DroEre_CAF1 72316 92 - 1 -----GGCAGCGGCAA-AAA---AGAAAGGUCAGGCGGCAAGGGCUCAAAGCAAACUGUUUGCCUUGUGCCUG-GGCAAAUAUUUGCAACUCGCC-----GCCUUUU -----....(((((..-...---.......(((((((.(((((.....((((.....)))).)))))))))))-)((((....)))).....)))-----))..... ( -31.70) >DroYak_CAF1 68243 91 - 1 -----GGCAGCAA-AA-AAA---GAAAAGGUCAACUGGCAAGGGCUUUAAGCUAACAGUUUGCCUUGUGCCUG-GCCAAAUAUUUGCAAUUCGCU-----GCCUUUU -----((((((..-..-(((---.....(((((...(((((((((...((((.....))))))))).))))))-))).....))).......)))-----))).... ( -28.90) >DroAna_CAF1 93632 94 - 1 -----AGCAGCGUCGAAAAA---GGGAAGGUCAAGUGGCAAAGGCUUAAAGCUAACUGUUUGGUCUGUGCCCGGGGCAAAUAUUUGCAAUUUUCU-----GCACUGU -----.((((.((.(((((.---(((.((..((((..(....(((.....)))..)..))))..))...)))...((((....))))..))))).-----)).)))) ( -30.30) >consensus _____GGCAGCGGCAA_AAA___AGAAAGGUCAAGUGGCAAGGGCUUAAAGCAAACUGUUUGCCUUGUGCCUG_GGCAAAUAUUUGCAAUUCGCU_____GCCUUUU ........................(((((((..((((((....)).....(((((.((((((((..........)))))))))))))....)))).....))))))) (-18.05 = -18.33 + 0.28)

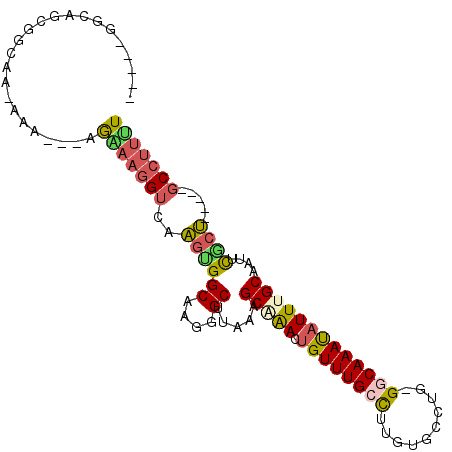
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:10 2006