| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,719,173 – 11,719,263 |
| Length | 90 |
| Max. P | 0.725265 |

| Location | 11,719,173 – 11,719,263 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -16.45 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11719173 90 - 20766785 -UCUGCAU-----UU-GCAUUUUCAGGGUCGUCGAAAACGGCAAGGAAACUGUCUUUUCAUAUGAAAAUGAUGUUCUGAAAUCAAAGACUGUGAUGG------------- -..(((..-----..-)))...(((.((((...(((((.((((.(....)))))))))).........((((........))))..)))).)))...------------- ( -22.70) >DroVir_CAF1 4974 110 - 1 UGUCUCAUGAUCUUUCUAUAUUGCAGCGUCGUCGAAAAUGGCAAGGAAACUGUCUUUUCAUAUGAAAAUGAUGUCCUUAAAUCAAAGACUGUGAACGGAGUAUCCCAAAA .((.((((..(((((..........(((((...(((((.((((.(....))))))))))..((....))))))).........)))))..))))))((......)).... ( -21.71) >DroGri_CAF1 5289 107 - 1 AGUUUCUUCUAC---UCUUUUUGCAGUGUCGUCGAAAACGGCAAGGAAACUGUCUUUUCAUAUGAAAAUGAUGUCCUUAAAUCAAAGACUGUGAAUGGAGUAUCCCAAAA .........(((---(((.((..(((((((((.....)))))(((((....((((((((....))))).))).))))).........))))..)).))))))........ ( -29.70) >DroEre_CAF1 4558 102 - 1 AUUUGCAU-----UU-GC--UUUCAGGGUUGUCGAAAACGGCAAGGAAACUGUCUUUUCAUAUGAAAAUGAUGUCCUUAAAUCAAAGACUGUGAACGGAGUGUCUCAAAA ....((((-----((-.(--.((((.((((((((....))))(((((....((((((((....))))).))).)))))........)))).)))).)))))))....... ( -24.20) >DroYak_CAF1 4553 102 - 1 AUGUGCAU-----UU-AC--UUGCAGAGUUGUCGAAAACGGCAAGGAAACUGUCUUUUCAUAUGAAAAUGAUGUCCUUAAAUCAAAGACUGUGAACGGAGUGUCCCAAAA ..(..(((-----((-..--...((.((((((((....))))(((((....((((((((....))))).))).)))))........)))).))....)))))..)..... ( -23.30) >DroMoj_CAF1 5069 97 - 1 -------------UUUCCCCUUGCAGCGUCGUCGAAAAUGGCAAGGAAACUGUCUUUUCAUAUGAGAACGAUGUCCUUAAAUCAAAGACUGUGAAUGGAGUAUCCCAAAA -------------.((((..(..(((((((((.....)))))(((((....((((((((....))))).))).)))))........).)))..)..)))).......... ( -22.20) >consensus AUUUGCAU_____UU_CC__UUGCAGCGUCGUCGAAAACGGCAAGGAAACUGUCUUUUCAUAUGAAAAUGAUGUCCUUAAAUCAAAGACUGUGAACGGAGUAUCCCAAAA ....................(..(((.(((((.....)))))(((((....((((((((....))))).))).)))))..........)))..)................ (-16.45 = -15.90 + -0.55)

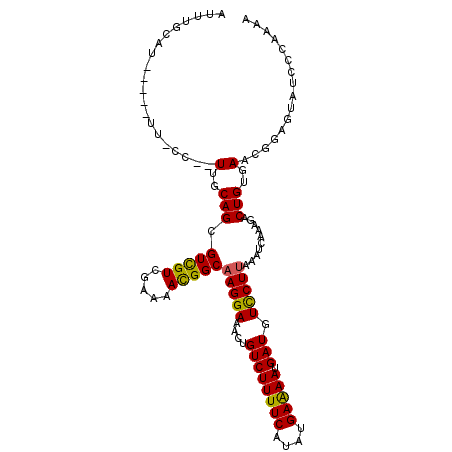

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:54 2006