
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,702,785 – 11,703,001 |
| Length | 216 |
| Max. P | 0.869221 |

| Location | 11,702,785 – 11,702,904 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -23.73 |
| Energy contribution | -23.54 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
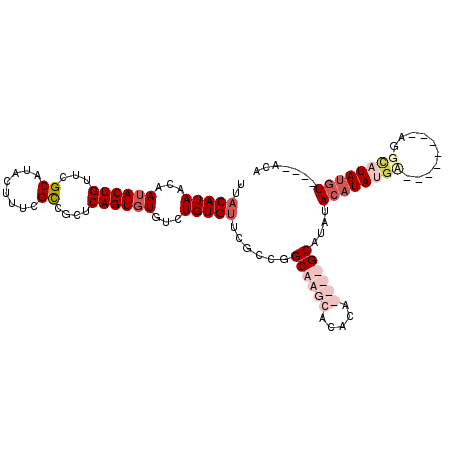
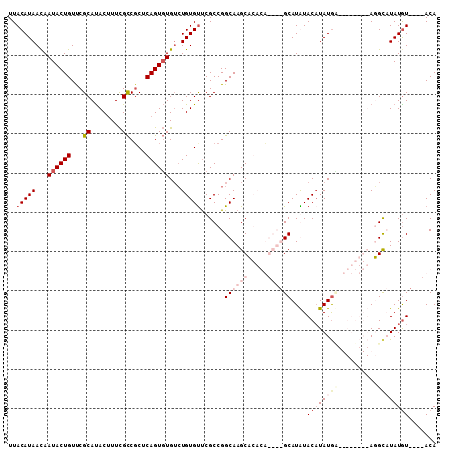
>2R_DroMel_CAF1 11702785 119 - 20766785 UACAU-UUACAACAAAACAGUAGAAAAACAAACAGAAAGAGAAAAGCGAGCUGAUAAGGCCAAAGGGGCCGGGUUCGUUCGGAGUCCGUACGAACGAACAGUCCACCGAAUAAUCCAAUC ....(-((((.........))))).......................(..(((....((((.....))))..((((((.(((...))).))))))...)))..)................ ( -24.70) >DroSim_CAF1 998 115 - 1 UACAU-UUACAACAAAACAGCAGAAAAACAAACAGAAAGAGACAAGCGAGCUGAUAAGGCCAAAGGGGCCGGGUUCGUUCGGAGUCCGUACG----AACAGUCCACCGAAUAAUCCAAUC .....-...........((((............................))))....((((.....))))(((((.((((((.(..(.....----....)..).))))))))))).... ( -23.89) >DroEre_CAF1 1004 114 - 1 UCGAU-UUACAACAAAACAGCAGAAAAACAAACAGAAGGGGC-AAGCGAGCUGAUAAGGCCAAAGGGGCCGGGUUCGUUCGGAGUCCGUACG----AACAGUCCGCCGAAUAAUCCAAUC .....-...............................(((..-...((.((.(((..((((.....))))..((((((.(((...))).)))----))).))).)))).....))).... ( -28.70) >DroYak_CAF1 1005 115 - 1 CUGUUUUUACAACAAAACAGCAGAAAAACAAACAGAAGGAGA-CAGCGGGCUGAUAAGGCCAAAGGGGCCGGGUUCGUUCGGAGUCCGUACG----AACAGUCCGCCGAAUAAUCCAAUC (((((((......))))))).................(((..-(.(((((((.....((((.....))))..((((((.(((...))).)))----)))))))))).).....))).... ( -37.60) >consensus UACAU_UUACAACAAAACAGCAGAAAAACAAACAGAAAGAGA_AAGCGAGCUGAUAAGGCCAAAGGGGCCGGGUUCGUUCGGAGUCCGUACG____AACAGUCCACCGAAUAAUCCAAUC .................((((............................))))....((((.....))))(((((.((((((.(..(((........)).)..).))))))))))).... (-23.73 = -23.54 + -0.19)

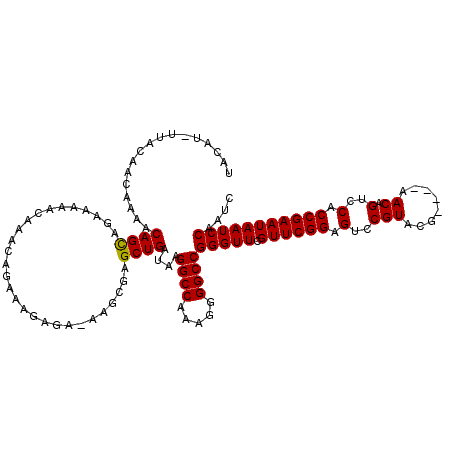
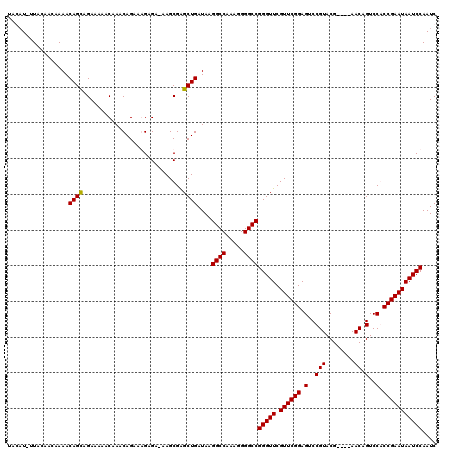
| Location | 11,702,865 – 11,702,968 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -12.88 |
| Energy contribution | -14.88 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11702865 103 + 20766785 UCUUUCUGUUUGUUUUUCUACUGUUUUGUUGUAA-AUGUAUGU----ACAUAUGCCU--------UUAUAUGUAUAUGCAAGCACACAGCUUGCCGGCGAACACAGACAAACUGAG ....((.((((((.......(((((((((((...-....((((----(((((((...--------.)))))))))))((((((.....))))))))))))).)))))))))).)). ( -30.71) >DroSim_CAF1 1074 107 + 1 UCUUUCUGUUUGUUUUUCUGCUGUUUUGUUGUAA-AUGUAUGUAUUUACAUAUACCU--------UCAUAUGUAUAUGCAAGCACACAGCUUGCCGGCGAACACAGACACACUGAG ((....(((((((..(((.((((.(..(((((..-.(((.(((((.(((((((....--------..))))))).))))).))).)))))..).))))))).)))))))....)). ( -30.00) >DroEre_CAF1 1079 107 + 1 CCCUUCUGUUUGUUUUUCUGCUGUUUUGUUGUAA-AUCGAUGU----AUAUAGGCAUACACACAUGCAUAAGUACAUGC----UGUGUGCUUGCCGGCGGACACAGACACACUGAG ......(((((((...((((((.....((((...-..))))..----.....((((..((((((.((((......))))----))))))..)))))))))).)))))))....... ( -34.60) >DroYak_CAF1 1080 108 + 1 UCCUUCUGUUUGUUUUUCUGCUGUUUUGUUGUAAAAACAGUGU----ACAUAGGCAUACACACAAACAUAUGUACAUGC----UGUGUGCUUGCCGGCGAGCACAUCCACACUGAG ......((((((((.....((((((((......))))))))..----.....((((..((((((..(((......))).----))))))..))))))))))))............. ( -29.90) >consensus UCCUUCUGUUUGUUUUUCUGCUGUUUUGUUGUAA_AUCUAUGU____ACAUAGGCAU________UCAUAUGUACAUGC____ACACAGCUUGCCGGCGAACACAGACACACUGAG ......(((((((..(((.((((..............((((((....))))))........................((((((.....))))))))))))).)))))))....... (-12.88 = -14.88 + 2.00)

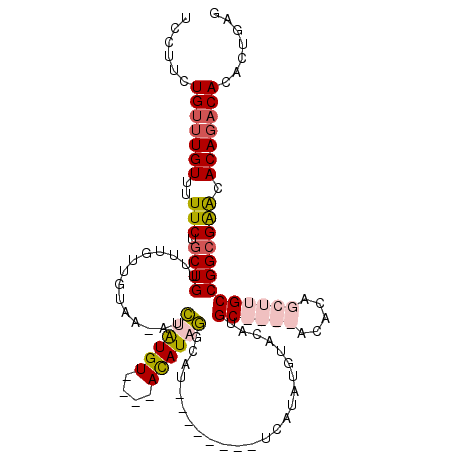
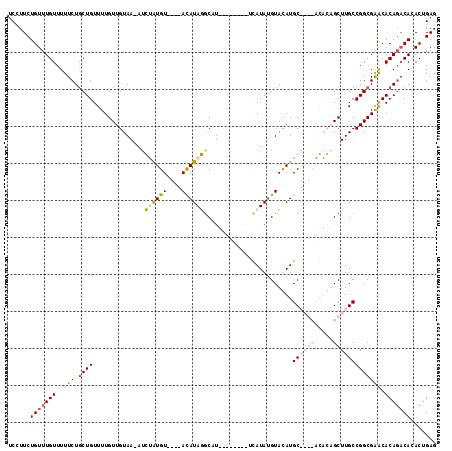
| Location | 11,702,904 – 11,703,001 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -11.27 |
| Energy contribution | -15.15 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.38 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11702904 97 - 20766785 UUACAUAACAAUACUGUCCGCACACUUUUGUCGCUCAGUUUGUCUGUGUUCGCCGGCAAGCUGUGUGCUUGCAUAUACAUAUAA--------AGGCAUAUGU----ACA .((((((((((.((((..((.(((....)))))..))))))))........(((.((((((.....)))))).(((....))).--------.))).)))))----).. ( -26.40) >DroSim_CAF1 1113 101 - 1 UUACAUAAUAAUACUGUUCGCAUACUUUCGCCGCUCAGUGUGUCUGUGUUCGCCGGCAAGCUGUGUGCUUGCAUAUACAUAUGA--------AGGUAUAUGUAAAUACA ..(((((...((((((..(((........).))..))))))...)))))......((((((.....))))))...((((((((.--------...))))))))...... ( -26.00) >DroEre_CAF1 1118 101 - 1 UUACAUAACAAUACUGUUCGCAUACUUUCGCCUCUCAGUGUGUCUGUGUCCGCCGGCAAGCACACA----GCAUGUACUUAUGCAUGUGUGUAUGCCUAUAU----ACA ..(((((...((((((...((........))....))))))...))))).....((((.(((((((----(((((....))))).))))))).)))).....----... ( -33.50) >DroYak_CAF1 1120 101 - 1 UUACAUAACAAUACUGUUCGCAUACGUUCGCCGCUCAGUGUGGAUGUGCUCGCCGGCAAGCACACA----GCAUGUACAUAUGUUUGUGUGUAUGCCUAUGU----ACA .((((((...........((..((((((((((.....).)))))))))..))..((((.(((((((----(((((....)))).)))))))).)))))))))----).. ( -34.00) >consensus UUACAUAACAAUACUGUUCGCAUACUUUCGCCGCUCAGUGUGUCUGUGUUCGCCGGCAAGCACACA____GCAUAUACAUAUGA________AGGCAUAUGU____ACA ..(((((...((((((...((........))....))))))...)))))......((((((.....))))))....((((((((..........))))))))....... (-11.27 = -15.15 + 3.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:50 2006