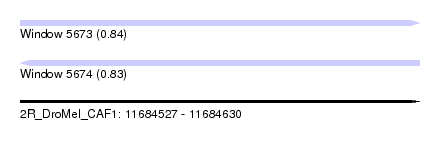
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,684,527 – 11,684,630 |
| Length | 103 |
| Max. P | 0.842729 |

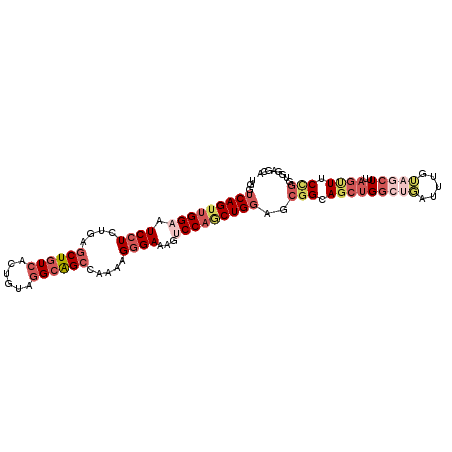
| Location | 11,684,527 – 11,684,630 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -24.34 |
| Energy contribution | -26.98 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
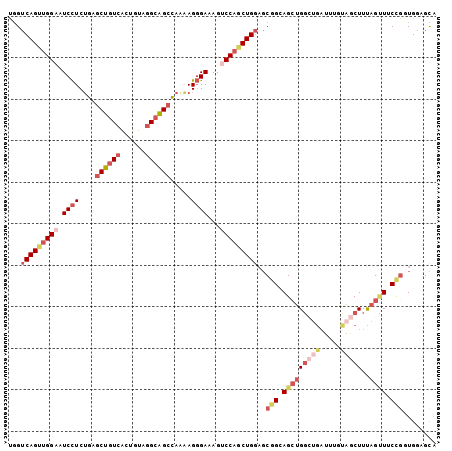
>2R_DroMel_CAF1 11684527 103 + 20766785 UGGUCAGUUGGAAUCCUCUGAGCUGUCACUGUAGGCAGCCAAGAGGGAAAGUCCAGCUGGAGCGGCAGCUGGCUGAUUUGUAGCUUUAGUUUCCGCUGGUGCA ...(((((((((.((((((..((((((......))))))..))))))....)))))))))(((((.(((((((((.....)))))..)))).)))))...... ( -47.10) >DroPse_CAF1 19471 100 + 1 GUGUCAGGUGGAAUCCUCUGAGCUAUCGCUAUAGGCGGCUAGAAGGGAUAGACCCGCUG---CUGGAGCUGGGGUCUUUGAAACUUCAGCUGCCUUUGGAAUA ...(((((.((...))))))).......(((.((((((((.((((....((((((.(.(---(....)).)))))))......)))))))))))).))).... ( -38.40) >DroSec_CAF1 24707 103 + 1 UGGUCAGUUGGAAUCCUCUGAGCUGUCACUGUAGGCAGCCAAAAGGGAAAGUCCAGCUGGAGCGGCAGCUGGCUGAUUUGUAGCUUUAGUUUCCGGUGGAGCA ...(((((((((.((((.((.((((((......))))))))...))))...)))))))))(.(((.(((((((((.....)))))..)))).))).)...... ( -38.00) >DroSim_CAF1 20608 103 + 1 UGGUCAGUUGGAAUCCUCUGAGCUGUCACUGUAGGCAGCCAAAAGGGAAAGUCCAGCUGGAGCGGCAGCUGGCUGAUUUGUAGCUUUAGUUUCCGGUGGAGCA ...(((((((((.((((.((.((((((......))))))))...))))...)))))))))(.(((.(((((((((.....)))))..)))).))).)...... ( -38.00) >DroEre_CAF1 20671 103 + 1 UGGUCAGUUGGAAUCCUCUGAGCUGUCACUGUAGGCAGCCAAGAGGGAUAGUCCAACUGGAGCGGCAGCUGGCUGAUUUGUAGCUUUAGUUUCCGGUGGUGCG ...(((((((((.((((((..((((((......))))))..))))))....)))))))))(.(((.(((((((((.....)))))..)))).))).)...... ( -42.60) >DroAna_CAF1 22684 103 + 1 UGGUCAGUUGGAAUCUUCGGAACUGUCGCUGUAAGCAGCCAAAAGUGAUAGCCCAGCUGGCGCGGAAGAUGCAGGAGUAGGGGCUUUUGGUUCUGCUGCCGUA ..(((((((((..((....)).((((((((.............)))))))).)))))))))((((.....((((((.((((....)))).))))))..)))). ( -39.92) >consensus UGGUCAGUUGGAAUCCUCUGAGCUGUCACUGUAGGCAGCCAAAAGGGAAAGUCCAGCUGGAGCGGCAGCUGGCUGAUUUGUAGCUUUAGUUUCCGGUGGAGCA ...(((((((((.((((....((((((......)))))).....))))...)))))))))..(((.(((((((((.....)))))..)))).)))........ (-24.34 = -26.98 + 2.64)

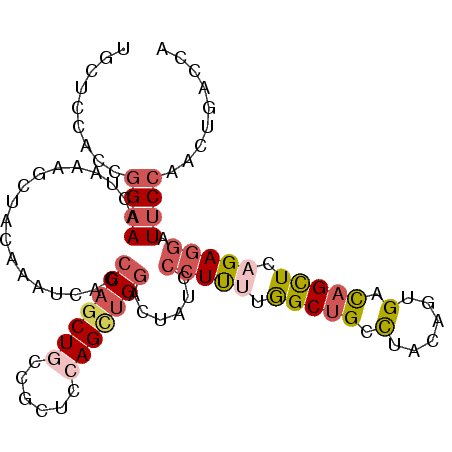
| Location | 11,684,527 – 11,684,630 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11684527 103 - 20766785 UGCACCAGCGGAAACUAAAGCUACAAAUCAGCCAGCUGCCGCUCCAGCUGGACUUUCCCUCUUGGCUGCCUACAGUGACAGCUCAGAGGAUUCCAACUGACCA .....(((.((((................(((((((((......))))))).))...(((((.(((((.(......).))))).))))).))))..))).... ( -33.40) >DroPse_CAF1 19471 100 - 1 UAUUCCAAAGGCAGCUGAAGUUUCAAAGACCCCAGCUCCAG---CAGCGGGUCUAUCCCUUCUAGCCGCCUAUAGCGAUAGCUCAGAGGAUUCCACCUGACAC ...(((..((((.(((((((......(((((((.((....)---).).))))))....)))).))).))))..(((....)))....)))............. ( -30.30) >DroSec_CAF1 24707 103 - 1 UGCUCCACCGGAAACUAAAGCUACAAAUCAGCCAGCUGCCGCUCCAGCUGGACUUUCCCUUUUGGCUGCCUACAGUGACAGCUCAGAGGAUUCCAACUGACCA .(((.....(....)...)))......(((((((((((......)))))))......(((((.(((((.(......).))))).))))).......))))... ( -30.30) >DroSim_CAF1 20608 103 - 1 UGCUCCACCGGAAACUAAAGCUACAAAUCAGCCAGCUGCCGCUCCAGCUGGACUUUCCCUUUUGGCUGCCUACAGUGACAGCUCAGAGGAUUCCAACUGACCA .(((.....(....)...)))......(((((((((((......)))))))......(((((.(((((.(......).))))).))))).......))))... ( -30.30) >DroEre_CAF1 20671 103 - 1 CGCACCACCGGAAACUAAAGCUACAAAUCAGCCAGCUGCCGCUCCAGUUGGACUAUCCCUCUUGGCUGCCUACAGUGACAGCUCAGAGGAUUCCAACUGACCA .((......(....)...((((...........))))...))..((((((((..((((.(((.(((((.(......).))))).))))))))))))))).... ( -31.90) >DroAna_CAF1 22684 103 - 1 UACGGCAGCAGAACCAAAAGCCCCUACUCCUGCAUCUUCCGCGCCAGCUGGGCUAUCACUUUUGGCUGCUUACAGCGACAGUUCCGAAGAUUCCAACUGACCA ...((.(((((..(((((((.(((..((..(((.......)))..))..)))......))))))))))))........(((((..((....)).))))).)). ( -22.90) >consensus UGCUCCACCGGAAACUAAAGCUACAAAUCAGCCAGCUGCCGCUCCAGCUGGACUAUCCCUUUUGGCUGCCUACAGUGACAGCUCAGAGGAUUCCAACUGACCA .........((((..................(((((((......)))))))......(((((.(((((.(......).))))).))))).))))......... (-20.32 = -21.43 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:33 2006