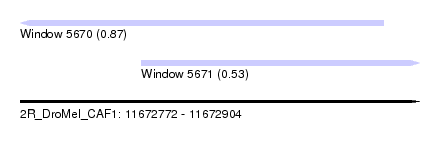
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,672,772 – 11,672,904 |
| Length | 132 |
| Max. P | 0.866816 |

| Location | 11,672,772 – 11,672,892 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -8.65 |
| Energy contribution | -10.05 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
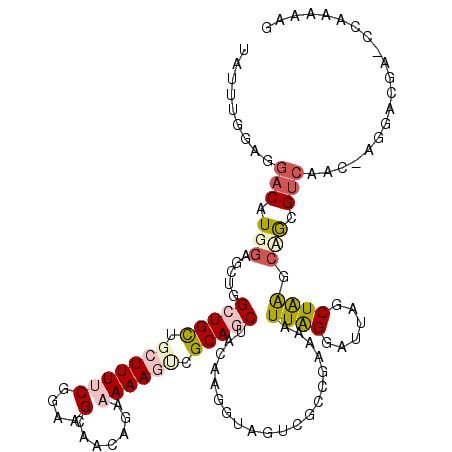
>2R_DroMel_CAF1 11672772 120 - 20766785 UAUUUGAAGGACAUGAAGCUGGCUGCUGCUUUUCGCAAGCAACAGAAAAAGUCGCAGCUACAAGUUAGUCGCCGAAAAUUAGGAUUAGCUAAGCAGCGUCAACUAGGACGAUCCAAAAAG ........((((.....(((((((((.(((((((....).......)))))).)))))....((((((((............))))))))...))))(((......)))).)))...... ( -31.61) >DroSec_CAF1 12832 120 - 1 UAUUUUGAGGACAUGGAAAUGGCUGCUGGUUUACGGAAGCACAAGAAAAAGUCGCAGCAACACGGUUAUCACCGAAAAUUAGGAUUAGCUAAGCAGCGUCAACCAGGACGAAUCAAAAAG ..((((((((.(((....))).)).((((((.(((...((((........)).)).((....((((....))))....((((......))))))..))).))))))......)))))).. ( -27.60) >DroSim_CAF1 8806 120 - 1 UAUUUUGAGGACAUGGAAAUGGCUGCUUGUUUGCCGAAGCAAAAGAAAAAGUCGCAGCAACACGGUUAUCGCCGAAAAUUAGGAUUAGCUAAGCUGCGUCAACCAGGACGAAUCAAAAAG ..(((((((..(.(((....((((.....(((((....)))))......))))(((((....((((....))))....((((......))))))))).....))))..)...)))))).. ( -31.10) >DroEre_CAF1 8642 108 - 1 UACUUGGAGAACAUGGAGCUGGGUGUUGCUUUUCGGAAGCAACAGCAAAAGCAGCAGCUCCAAAGUAGUCGCCGAAGAUUAGGUGGAGCUGGACCACGUCGAC------------UAGGA ..(((((.((...(((.(((.(.(((((((((...))))))))).)...)))..((((((((...(((((......)))))..))))))))..)))..))..)------------)))). ( -43.30) >DroYak_CAF1 8835 111 - 1 UACUUGGAGGACAUGGAGCUGGGUGCUACUUUUCGGAAGCAACAGCAAAAGCAGCAGCUUCUAAGUAGUCGCCGUAGUUUGGGUGUAGCUGGACCAAGACGAU---------CCCCAAAG ...((((.((((...((((((((((((((((...((((((..(.((....)).)..)))))))))))).)))).)))))).(((.(....).))).....).)---------)))))).. ( -35.80) >consensus UAUUUGGAGGACAUGGAGCUGGCUGCUGCUUUUCGGAAGCAACAGAAAAAGUCGCAGCUACAAGGUAGUCGCCGAAAAUUAGGAUUAGCUAAGCAGCGUCAAC_AGGACGA_CCAAAAAG .........(((.(((.....(((((.(((((((....).......)))))).)))))....................((((......)))).))).))).................... ( -8.65 = -10.05 + 1.40)

| Location | 11,672,812 – 11,672,904 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.48 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -7.88 |
| Energy contribution | -10.12 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11672812 92 + 20766785 AAUUUUCGGCGACUAACUUGUAGCUGCGACUUUUUCUGUUGCUUGCGAAAAGCAGCAGCCAGCUUCAUGUCCUUCAAAUAAUCGUCGACCUU---------------------------- .....(((((((.((((.((.(((((.(.......(((((((((.....))))))))))))))).)).))........)).)))))))....---------------------------- ( -26.01) >DroSec_CAF1 12872 92 + 1 AAUUUUCGGUGAUAACCGUGUUGCUGCGACUUUUUCUUGUGCUUCCGUAAACCAGCAGCCAUUUCCAUGUCCUCAAAAUAUUUGCUGACCUU---------------------------- ......((((....)))).(((((((.(((........))((....))...)))))))).........(((..(((.....)))..)))...---------------------------- ( -14.30) >DroSim_CAF1 8846 92 + 1 AAUUUUCGGCGAUAACCGUGUUGCUGCGACUUUUUCUUUUGCUUCGGCAAACAAGCAGCCAUUUCCAUGUCCUCAAAAUAUUUGCUAACCUU---------------------------- .(((((.((.((((.....((.(((((((.....)).(((((....)))))...))))).)).....)))))).))))).............---------------------------- ( -19.60) >DroEre_CAF1 8670 120 + 1 AAUCUUCGGCGACUACUUUGGAGCUGCUGCUUUUGCUGUUGCUUCCGAAAAGCAACACCCAGCUCCAUGUUCUCCAAGUAAUCGCUGACCUUGGAUGCCGAUUCGGGGCCUACUGUACUC .....(((((((.(((((((((((((..((....))((((((((.....))))))))..))))))))........))))).)))))))((((((((....))))))))............ ( -43.60) >DroYak_CAF1 8866 93 + 1 AAACUACGGCGACUACUUAGAAGCUGCUGCUUUUGCUGUUGCUUCCGAAAAGUAGCACCCAGCUCCAUGUCCUCCAAGUAAUC---------------------------CACUUUACUC .......((.(.((((((.(((((.((.((....)).)).)))))....))))))).)).................(((((..---------------------------....))))). ( -20.70) >consensus AAUUUUCGGCGACUACCUUGUAGCUGCGACUUUUUCUGUUGCUUCCGAAAAGCAGCAGCCAGCUCCAUGUCCUCCAAAUAAUCGCUGACCUU____________________________ .....(((((((..........((((..(((....(((((((((.....)))))))))..)))..)).))...........)))))))................................ ( -7.88 = -10.12 + 2.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:29 2006