
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,653,499 – 11,653,649 |
| Length | 150 |
| Max. P | 0.867539 |

| Location | 11,653,499 – 11,653,609 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -25.45 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
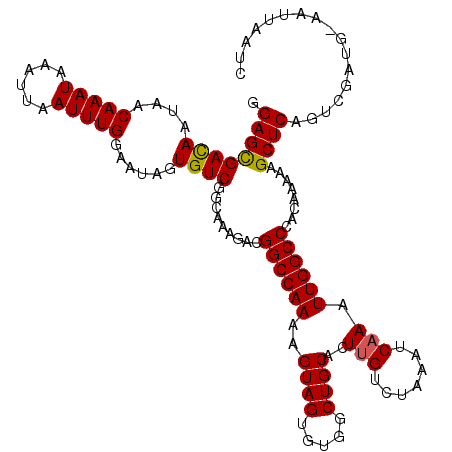
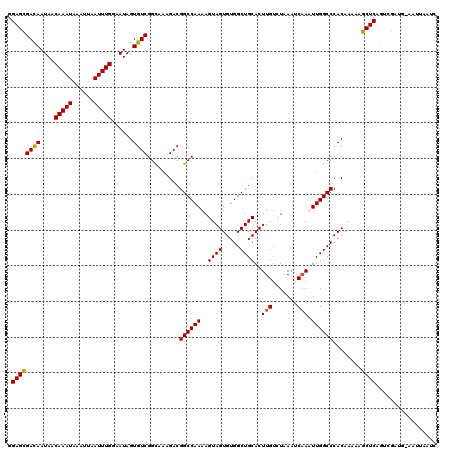
>2R_DroMel_CAF1 11653499 110 - 20766785 GGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCAAAGACGGCCAAAAGUAGUGUGGCUGCACUUGUCUAAAUCAAAUUGGCCCACAA-AAGCUCAGUCGAUG--------- .((((((((....(((((......)))))......)))).........((((((..((((.....))))..(((.......))).)))))).....-..))))........--------- ( -26.90) >DroSim_CAF1 26866 120 - 1 GGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGUGAAGACGGCCAAAAGUAGUGUGGCUGCACUUGUCUAAAUCAAAUUGGCCCACAAAAAGCUCAGUUGAUGCAAGUAAUC .((((((((....(((((......)))))......)))).(((.....((((((..((((.....))))..(((.......))).))))))))).....))))................. ( -27.80) >DroEre_CAF1 25771 118 - 1 GGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCA--GACGGCCAAAAGUAGUGUGGCUGCACUUGUCUGAAACAAUUUGGCCCACAAAAAGCUCAGUCGAUUAAAUUAAUC .((((((((....(((((......)))))......))))....--...(((((((.((((.....))))..((((.....)))))))))))........))))................. ( -28.80) >DroYak_CAF1 26369 120 - 1 GGAGUGAUAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCAACGACGGCCAAAAGUAGUGUGGCUGCACUUGGGUAAGCCCAUUUGGCCCACAAAAAGCUCGACCGAUUUAAUUCAUC ...................((((((.((((...(((((((....))))(((((((.((((.....))))...((((....)))))))))))........)))...)))).)))))).... ( -36.30) >consensus GGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCAAAGACGGCCAAAAGUAGUGUGGCUGCACUUGUCUAAAUCAAAUUGGCCCACAAAAAGCUCAGUCGAUG_AAUUAAUC .((((((((....(((((......)))))......)))).........((((((..((((.....))))..(((.......))).))))))........))))................. (-25.45 = -25.32 + -0.13)

| Location | 11,653,529 – 11,653,649 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
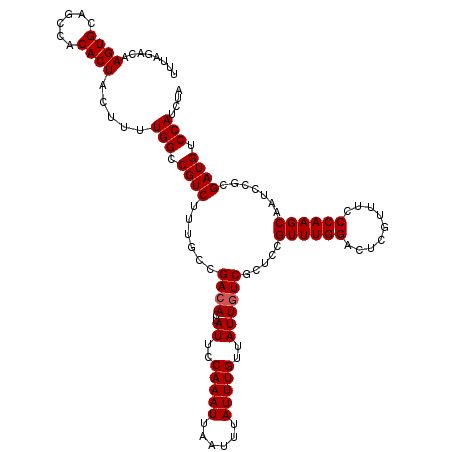
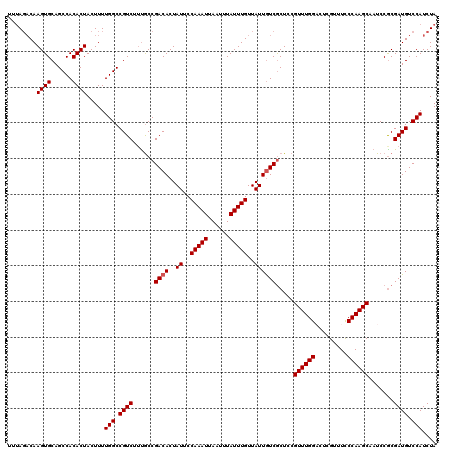
>2R_DroMel_CAF1 11653529 120 + 20766785 UUUAGACAAGUGCAGCCACACUACUUUUGGCCGUCUUUGCCGACACUAUUCCAAAUUAAUUUAUUUGUUAUUGUCGCUCCGUUUGGACUUGUUUCCCAAGCUAUCCACGAUGUCCAUCUA ..((((..((((......)))).....(((.((((...((.((((..((..(((((......)))))..))))))))...((((((.........)))))).......)))).))))))) ( -24.80) >DroSim_CAF1 26906 120 + 1 UUUAGACAAGUGCAGCCACACUACUUUUGGCCGUCUUCACCGACACUAUUCCAAAUUAAUUUAUUUGUUAUUGUCGCUCCGUUUGGACCUGUUUCCCAAGCAAUCCGCGAUGUCCAUCUA ..((((..((((......)))).....(((.((((.....(((((..((..(((((......)))))..)))))))....((((((.........)))))).......)))).))))))) ( -25.20) >DroEre_CAF1 25811 118 + 1 UUCAGACAAGUGCAGCCACACUACUUUUGGCCGUC--UGCCGACACUAUUCCAAAUUAAUUUAUUUGUUAUUGUCGCUCCGUUUGGUCUCGUUUUCCAAGCUAUCCGCGAUGUCCAUCUA ...(((..((((......)))).....(((.((((--.(((((((..((..(((((......)))))..)))))))....((((((.........)))))).....)))))).)))))). ( -25.80) >DroYak_CAF1 26409 120 + 1 CUUACCCAAGUGCAGCCACACUACUUUUGGCCGUCGUUGCCGACACUAUUCCAAAUUAAUUUAUUUGUUAUUAUCACUCCGUUUGGUCUCGUUUCCCAAGCAAUCCGCGAUGUCCAUCUA ........((((......)))).....(((.((((((....((....((..(((((......)))))..))..)).....((((((.........)))))).....)))))).))).... ( -21.50) >consensus UUUAGACAAGUGCAGCCACACUACUUUUGGCCGUCUUUGCCGACACUAUUCCAAAUUAAUUUAUUUGUUAUUGUCGCUCCGUUUGGACUCGUUUCCCAAGCAAUCCGCGAUGUCCAUCUA ........((((......)))).....(((.((((......((((..((..(((((......)))))..)))))).....((((((.........)))))).......)))).))).... (-21.02 = -21.27 + 0.25)

| Location | 11,653,529 – 11,653,649 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.62 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11653529 120 - 20766785 UAGAUGGACAUCGUGGAUAGCUUGGGAAACAAGUCCAAACGGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCAAAGACGGCCAAAAGUAGUGUGGCUGCACUUGUCUAAA ....((((((..(((....((...(....)...(((....))).(((((....(((((......)))))......))))))).....((((((.........))))))))).)))))).. ( -30.00) >DroSim_CAF1 26906 120 - 1 UAGAUGGACAUCGCGGAUUGCUUGGGAAACAGGUCCAAACGGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGUGAAGACGGCCAAAAGUAGUGUGGCUGCACUUGUCUAAA ....(((((((((((((((.....(....).)))))........(((((....(((((......)))))......))))))))).(.((((((.........)))))))...)))))).. ( -33.40) >DroEre_CAF1 25811 118 - 1 UAGAUGGACAUCGCGGAUAGCUUGGAAAACGAGACCAAACGGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCA--GACGGCCAAAAGUAGUGUGGCUGCACUUGUCUGAA .....(((((..((((....((((.....)))).(((.((...((((((....(((((......)))))......))))(((.--....)))....)).)).)))))))...)))))... ( -29.00) >DroYak_CAF1 26409 120 - 1 UAGAUGGACAUCGCGGAUUGCUUGGGAAACGAGACCAAACGGAGUGAUAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCAACGACGGCCAAAAGUAGUGUGGCUGCACUUGGGUAAG ....(((...((((.....))...(....)))..)))..(.(((((.......(((((......))))).......((((....))))(((((.........))))).))))).)..... ( -27.80) >consensus UAGAUGGACAUCGCGGAUAGCUUGGGAAACAAGACCAAACGGAGCGACAAUAACAAAUAAAUUAAUUUGGAAUAGUGUCGGCAAAGACGGCCAAAAGUAGUGUGGCUGCACUUGUCUAAA ....((((((..((((....((((.....)))).(((.((.....((((....(((((......)))))......))))(((.......))).......)).)))))))...)))))).. (-26.31 = -25.62 + -0.69)

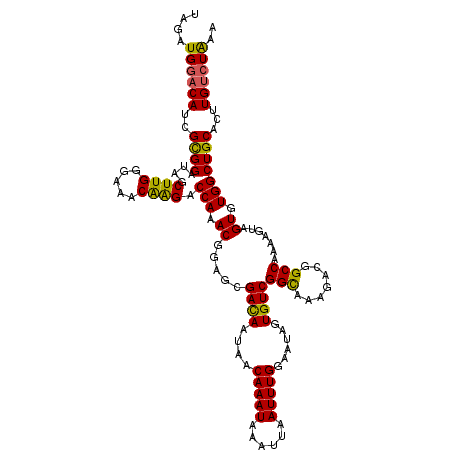
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:23 2006