| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,649,105 – 11,649,204 |
| Length | 99 |
| Max. P | 0.986176 |

| Location | 11,649,105 – 11,649,204 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
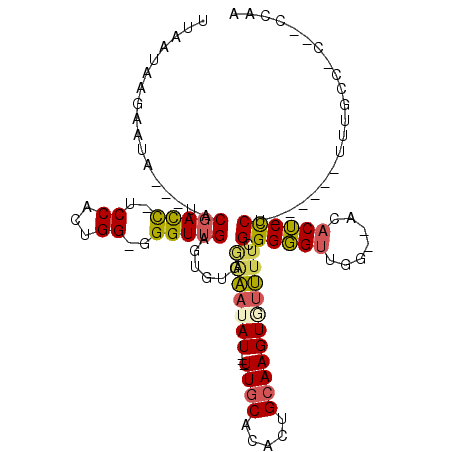
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
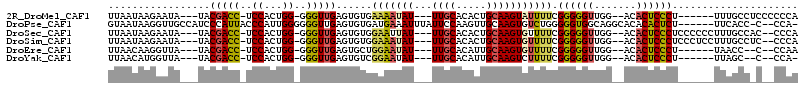
>2R_DroMel_CAF1 11649105 99 - 20766785 UUAAUAAGAAUA---UACGACC-UCCACUGG-GGGUUGAGUGUGAAAAUAU---UUGCACACUGCAAGUAUUUUCGGGGGUUGG--ACACUCCCU------UUUGCCUCCCCCCA .........(((---(.(((((-(((...))-)))))).))))((((((((---((((.....))))))))))))(((((..((--.((......------..))))..))))). ( -41.10) >DroPse_CAF1 24028 105 - 1 GUAAUAAGGUUGCCAUCCCAUUACCCAUUGGGGGGUUGAGUGUGAUGAAAUUUAUUCCAAGUUGCAAGUGUCUGGGGGUGGCAGGCACACACUCU------UUCACC-C--CCA- (((((..((.......)).)))))......((((((.((((((..((.(((((.....))))).)).(((((((.......))))))))))))).------...)))-)--)).- ( -32.80) >DroSec_CAF1 21548 103 - 1 UUAAUAAGAAUA---UACGACC-UCCACUGG-GGGUUGAGUGUGGAAUUAU---UUGCACACUGCAAGUGUUUUCGGGGGUUGG--ACACUCCCUCCCCCCUUUGCCAC--CCCA .......(((.(---(((((((-(((...))-))))).((((((.((....---)).))))))....)))).)))(((((..((--......))..)))))........--.... ( -36.80) >DroSim_CAF1 22477 103 - 1 UUAAUAAGAAUA---UACGACC-UCCACUGG-GGGUUGAGUGUGGAAAUAU---UUGCACACUGCAAGUGUUUUCGGGGGUUGG--ACACUCCCUCCCUCCUUUGCCUC--CCCA .........(((---(.(((((-(((...))-)))))).))))((((((((---((((.....))))))))))))((((((.((--(...........)))...)))))--)... ( -37.00) >DroEre_CAF1 21466 95 - 1 UUAACAAGGUUA---UACGACC-UCCACUGG-GGGUUGAGUGCUGGAAUAU---UUGCACAUUGCAAGUGUUUUCGGGGGUUGG--ACACUCCCU------UAACC--C--CCAA ......(((((.---...))))-)....(((-(((((((.....(((((((---((((.....))))))))))).((((((...--..)))))))------)))))--)--))). ( -38.30) >DroYak_CAF1 22023 94 - 1 UUAACAUGGUUA---UACGACC-UCCACUGG-GGGUUGAGUGUCGGAAUAU---UUGCACAUUGCAAGUCUUUUCGGGGGUUGG--ACACUCCCU------UUAGC--C--CCA- ............---..(((((-(((...))-)))))).....(((((.((---((((.....))))))..)))))((((((((--(.......)------)))))--)--)).- ( -32.10) >consensus UUAAUAAGAAUA___UACGACC_UCCACUGG_GGGUUGAGUGUGGAAAUAU___UUGCACACUGCAAGUGUUUUCGGGGGUUGG__ACACUCCCU______UUUGCC_C__CCAA .................(((((..((...))..)))))......(((((((...((((.....))))))))))).((((((.......))))))..................... (-13.18 = -13.18 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:18 2006