| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,642,523 – 11,642,639 |
| Length | 116 |
| Max. P | 0.986487 |

| Location | 11,642,523 – 11,642,639 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
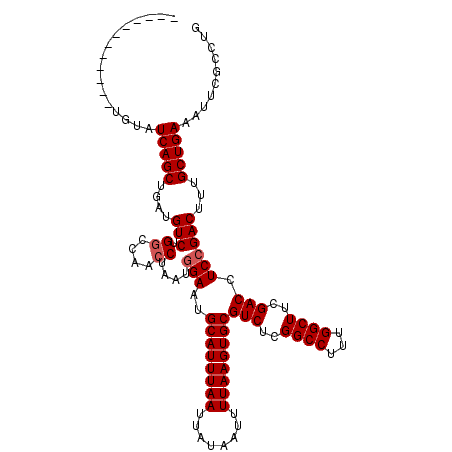
>2R_DroMel_CAF1 11642523 116 + 20766785 CACAUAGACGUGUGUAUCAGCUGAUGUCUGCCCAACCUAAUGGAAUGCAUUUAAUUAUAAUUUUAAGUGCGUCUCGGCCUUUGGCUUCGACCUCCGACUUUGCUGAAAUUCGCCUG (((((....)))))..(((((....(((...(((......)))...((((((((........))))))))(((..((((...))))..)))....)))...))))).......... ( -29.70) >DroSec_CAF1 14900 114 + 1 CACAUAGACG--UGUAUCAGCUGAUGUCUGGCCAACCUAAUGGAAUGCAUUUAAUUAUAAUUUUAAGUGCGUCUCGGCCUUUGGCUUCGACCUCCGACUUUGCUGAAAUUCGCCUG ....(((.((--.((.(((((....(((.((((..((....)).((((((((((........))))))))))...))))...((......))...)))...))))).)).)).))) ( -32.60) >DroSim_CAF1 15159 104 + 1 ------------UGUAUCAGCUGAUGUCUGGCCAACCUAAUGGAAUGCAUUUAAUUAUAAUUUUAAGUGCGUCUCGGCCUUUGGCUGCGACCUCCGACUUUGCUGAAAUUCGCCUG ------------....(((((....(((.((....))....(((..((((((((........))))))))(((.(((((...))))).))).))))))...))))).......... ( -32.00) >DroEre_CAF1 14963 103 + 1 -------------AUGUCAGCUGAUGUCUGGCCAACCUAAUUGAGUGCAUUUAAUUAUAAUUUUAAGUGCGUCUCGGCCUUUGGCUUCGACCUCCGACUUUGCUGAAUUUUGCAUG -------------...(((((.((.(((.((((((.....((((((((((((((........))))))))).)))))...))))))..))).)).......))))).......... ( -29.00) >DroYak_CAF1 15215 106 + 1 ----------UUUGUAUCAGCUGAUGUCUGGUUAGCCUAAUGGAAUGCAUUUAAUUAUAAUUUUAAGUGCGUCUCGGCCUUUGGCUUCGACCUCCGACUUUGCUGAAUUUCGCAUG ----------......(((((....(((.((((((((...((..((((((((((........))))))))))..))......))))..))))...)))...))))).......... ( -32.30) >consensus ____________UGUAUCAGCUGAUGUCUGGCCAACCUAAUGGAAUGCAUUUAAUUAUAAUUUUAAGUGCGUCUCGGCCUUUGGCUUCGACCUCCGACUUUGCUGAAAUUCGCCUG ................(((((....(((.((....))....(((..((((((((........))))))))(((..((((...))))..))).))))))...))))).......... (-26.34 = -26.74 + 0.40)


| Location | 11,642,523 – 11,642,639 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
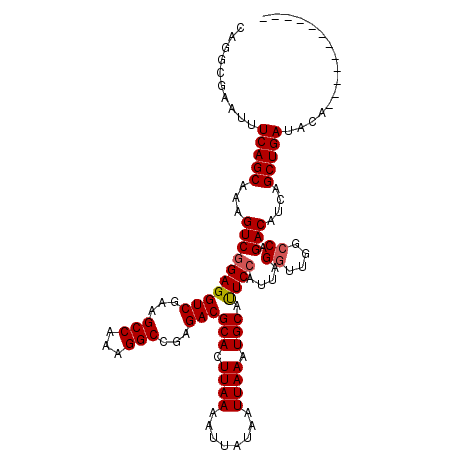
>2R_DroMel_CAF1 11642523 116 - 20766785 CAGGCGAAUUUCAGCAAAGUCGGAGGUCGAAGCCAAAGGCCGAGACGCACUUAAAAUUAUAAUUAAAUGCAUUCCAUUAGGUUGGGCAGACAUCAGCUGAUACACACGUCUAUGUG .(((((....(((((...(((...((((.........))))..)))(((.((((........)))).)))..((((......)))).........)))))......)))))..... ( -28.30) >DroSec_CAF1 14900 114 - 1 CAGGCGAAUUUCAGCAAAGUCGGAGGUCGAAGCCAAAGGCCGAGACGCACUUAAAAUUAUAAUUAAAUGCAUUCCAUUAGGUUGGCCAGACAUCAGCUGAUACA--CGUCUAUGUG .(((((....(((((...(((...(((....)))...((((((...(((.((((........)))).)))...((....)))))))).)))....)))))....--)))))..... ( -32.20) >DroSim_CAF1 15159 104 - 1 CAGGCGAAUUUCAGCAAAGUCGGAGGUCGCAGCCAAAGGCCGAGACGCACUUAAAAUUAUAAUUAAAUGCAUUCCAUUAGGUUGGCCAGACAUCAGCUGAUACA------------ ..........(((((...((((((((((.(.(((...))).).)))(((.((((........)))).))).))))....((....)).)))....)))))....------------ ( -26.20) >DroEre_CAF1 14963 103 - 1 CAUGCAAAAUUCAGCAAAGUCGGAGGUCGAAGCCAAAGGCCGAGACGCACUUAAAAUUAUAAUUAAAUGCACUCAAUUAGGUUGGCCAGACAUCAGCUGACAU------------- ..........(((((...(((.((((((...(((...)))...)))(((.((((........)))).))).))).....((....)).)))....)))))...------------- ( -24.60) >DroYak_CAF1 15215 106 - 1 CAUGCGAAAUUCAGCAAAGUCGGAGGUCGAAGCCAAAGGCCGAGACGCACUUAAAAUUAUAAUUAAAUGCAUUCCAUUAGGCUAACCAGACAUCAGCUGAUACAAA---------- ..........(((((...((((((((((...(((...)))...)))(((.((((........)))).))).))))....((....)).)))....)))))......---------- ( -24.50) >consensus CAGGCGAAUUUCAGCAAAGUCGGAGGUCGAAGCCAAAGGCCGAGACGCACUUAAAAUUAUAAUUAAAUGCAUUCCAUUAGGUUGGCCAGACAUCAGCUGAUACA____________ ..........(((((...((((((((((...(((...)))...)))(((.((((........)))).))).))))....((....)).)))....)))))................ (-23.40 = -23.64 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:17 2006