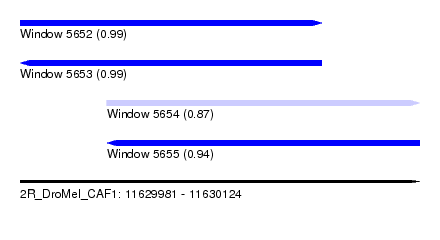
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,629,981 – 11,630,124 |
| Length | 143 |
| Max. P | 0.992546 |

| Location | 11,629,981 – 11,630,089 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -24.03 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11629981 108 + 20766785 CGCUCAUUUCAAGAGACCCAUAUAUGAUUGGGAUGAACCCACGACCAGUUACUCCAAGAACUACACACCCACUUGGAGUAAACUUUGAUCGCGGGCUCAUCCUUGGAA ......(((((((.((((((........)))).(((.(((.(((.(((((((((((((.............))))))))))...))).))).))).)))))))))))) ( -36.62) >DroPse_CAF1 1388 101 + 1 CCCUCUUGUCAAA----GCUU-UUCAAUUGGGCGCAACCAAAGGCGGCCUCCUCCAGCCA-GACACACCCACUGG-AGGGGUCCUCCUCGAUGGGAUGUUCCCCUGAA .............----....-((((...(((.(((.(((.(((.((.(((((((((...-..........))))-))))).)).)))...)))..))).))).)))) ( -32.82) >DroSec_CAF1 2512 106 + 1 CGCUCUUUUCAGGAGACACAU--AUGGUUUGGAUGAACCCACGACCACUUACUCCAAAAACUACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAA .(((((.....)))).)....--.......((((((.(((.((((((....((((((...............)))))).......)))))).))).))))))...... ( -34.66) >DroSim_CAF1 2528 106 + 1 CGCUCUUAUCAGGAGACGCAU--AUUGUUGGGAUGAACCCACGACCAGUUACUCCAAAAACUACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAU .((((((.....)))).))..--......(((((((.(((.(((((((((.((((((...............))))))..)))..)))))).))).)))))))..... ( -38.36) >DroEre_CAF1 2488 106 + 1 CGCUCUUUUCAGGAGAUGUAU--AUGGUUGGGAUGAACCCACGACGAGCUACUCCAAAAUUUACACACCCACUAGGAGGAAGCUUUGGUCGCGGGCUCAUCCUUAGAA ..((((.....))))......--......(((((((.(((.(((((((((.((((...................))))..)))))..)))).))).)))))))..... ( -34.91) >DroYak_CAF1 2516 106 + 1 CGCUCUUUUCAGGAAAUGCAU--UUGACUGGGAUGAACCCACGAUCAGCUACUCCAAAAACUACACACCCACCAGGAGGUAGCUUUGGUCGCGGGCUCAUCCUUGGAA .((..(((....)))..))..--....(.(((((((.(((.((((((((((((((...................))).)))))..)))))).))).))))))).)... ( -35.41) >consensus CGCUCUUUUCAGGAGACGCAU__AUGAUUGGGAUGAACCCACGACCAGCUACUCCAAAAACUACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAA .............................(((((((.(((.(((((((...((((...................))))......))))))).))).)))))))..... (-24.03 = -24.34 + 0.31)

| Location | 11,629,981 – 11,630,089 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.99 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11629981 108 - 20766785 UUCCAAGGAUGAGCCCGCGAUCAAAGUUUACUCCAAGUGGGUGUGUAGUUCUUGGAGUAACUGGUCGUGGGUUCAUCCCAAUCAUAUAUGGGUCUCUUGAAAUGAGCG ......(((((((((((((((((....((((((((((.............)))))))))).))))))))))))))))).............(.(((.......)))). ( -44.92) >DroPse_CAF1 1388 101 - 1 UUCAGGGGAACAUCCCAUCGAGGAGGACCCCU-CCAGUGGGUGUGUC-UGGCUGGAGGAGGCCGCCUUUGGUUGCGCCCAAUUGAA-AAGC----UUUGACAAGAGGG ((((.(((..((..(((..((((.((.(((((-(((((.((.....)-).)))))))).)))).))))))).))..)))...))))-...(----(((....)))).. ( -41.30) >DroSec_CAF1 2512 106 - 1 UUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGUAGUUUUUGGAGUAAGUGGUCGUGGGUUCAUCCAAACCAU--AUGUGUCUCCUGAAAAGAGCG ......(((((((((((((((((.......(((((((.............)))))))....))))))))))))))))).......--....(.(((.(....))))). ( -42.62) >DroSim_CAF1 2528 106 - 1 AUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGUAGUUUUUGGAGUAACUGGUCGUGGGUUCAUCCCAACAAU--AUGCGUCUCCUGAUAAGAGCG ......(((((((((((((((((..(((..(((((((.............))))))).)))))))))))))))))))).......--..((.(((.......))))). ( -43.52) >DroEre_CAF1 2488 106 - 1 UUCUAAGGAUGAGCCCGCGACCAAAGCUUCCUCCUAGUGGGUGUGUAAAUUUUGGAGUAGCUCGUCGUGGGUUCAUCCCAACCAU--AUACAUCUCCUGAAAAGAGCG ......(((((((((((((((...((((..((((....((((......)))).)))).)))).))))))))))))))).......--.........((....)).... ( -39.00) >DroYak_CAF1 2516 106 - 1 UUCCAAGGAUGAGCCCGCGACCAAAGCUACCUCCUGGUGGGUGUGUAGUUUUUGGAGUAGCUGAUCGUGGGUUCAUCCCAGUCAA--AUGCAUUUCCUGAAAAGAGCG ......((((((((((((((....((((((.(((...................)))))))))..)))))))))))))).......--.........((....)).... ( -38.11) >consensus UUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGUAGUUUUUGGAGUAACUGGUCGUGGGUUCAUCCCAACCAU__AUGCGUCUCCUGAAAAGAGCG ......(((((((((((((((((.......((((...................))))....))))))))))))))))).............................. (-28.60 = -28.99 + 0.39)


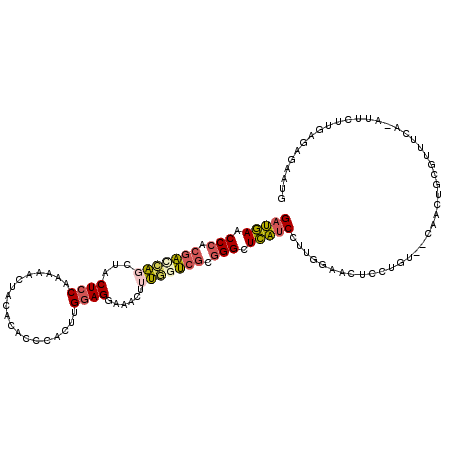
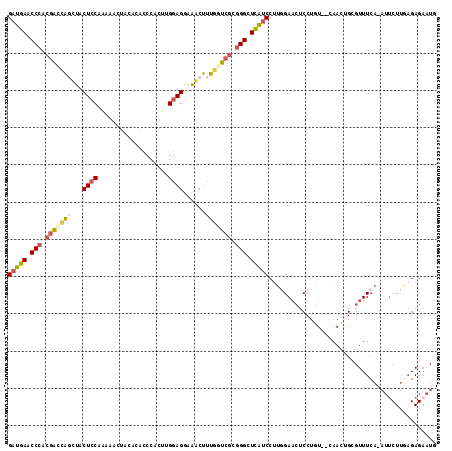
| Location | 11,630,012 – 11,630,124 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11630012 112 + 20766785 GAUGAACCCACGACCAGUUACUCCAAGAACUACACACCCACUUGGAGUAAACUUUGAUCGCGGGCUCAUCCUUGGAACUCCUUU--CAACAGCGUUUCA-AUUCUUGAGAGAAUG (((((.(((.(((.(((((((((((((.............))))))))))...))).))).))).)))))..((((((..((..--....)).))))))-(((((....))))). ( -33.82) >DroPse_CAF1 1414 113 + 1 GCGCAACCAAAGGCGGCCUCCUCCAGCCA-GACACACCCACUGG-AGGGGUCCUCCUCGAUGGGAUGUUCCCCUGAAGUCCUCUGAAAAUUGUGUUUCCUCUUCUAAUGAGCUUC ((((((.....((.((.(((((((((...-..........))))-))))).)).))(((..(((((.(((....)))))))).)))...))))))...(((.......))).... ( -31.82) >DroSec_CAF1 2541 112 + 1 GAUGAACCCACGACCACUUACUCCAAAAACUACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAACUCCUGU--CAACUGCGUUUCA-AUUCUUGAGAGAAUG (((((.(((.((((((....((((((...............)))))).......)))))).))).)))))..((((((.(..(.--...).).))))))-(((((....))))). ( -33.16) >DroSim_CAF1 2557 112 + 1 GAUGAACCCACGACCAGUUACUCCAAAAACUACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAUCUCCUGU--CAACUGCGUUUCA-AUUCUUGAGAGAAUG (((((.(((.(((((((((.((((((...............))))))..)))..)))))).))).)))))...((....))...--.............-(((((....))))). ( -32.36) >DroEre_CAF1 2517 112 + 1 GAUGAACCCACGACGAGCUACUCCAAAAUUUACACACCCACUAGGAGGAAGCUUUGGUCGCGGGCUCAUCCUUAGAACUCCUGU--CAACUGCGUUUCA-AUUCUUGAGAGAAUG (((((.(((.(((((((((.((((...................))))..)))))..)))).))).)))))..........((.(--(((..........-....)))).)).... ( -30.25) >DroYak_CAF1 2545 112 + 1 GAUGAACCCACGAUCAGCUACUCCAAAAACUACACACCCACCAGGAGGUAGCUUUGGUCGCGGGCUCAUCCUUGGAACUCCUGU--CAACAGCGUUUCA-AUACUUAAGAGAAUG (((((.(((.((((((((((((((...................))).)))))..)))))).))).))))).(((((((..(((.--...))).))))))-).............. ( -36.21) >consensus GAUGAACCCACGACCAGCUACUCCAAAAACUACACACCCACUUGGAGGAAACUUUGGUCGCGGGCUCAUCCUUGGAACUCCUGU__CAACUGCGUUUCA_AUUCUUGAGAGAAUG (((((.(((.(((((((...((((...................))))......))))))).))).)))))............................................. (-19.72 = -19.98 + 0.26)

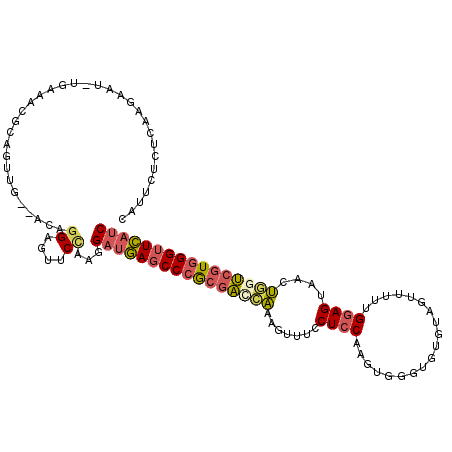
| Location | 11,630,012 – 11,630,124 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.89 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11630012 112 - 20766785 CAUUCUCUCAAGAAU-UGAAACGCUGUUG--AAAGGAGUUCCAAGGAUGAGCCCGCGAUCAAAGUUUACUCCAAGUGGGUGUGUAGUUCUUGGAGUAACUGGUCGUGGGUUCAUC ..((((.((((.(..-........).)))--).))))........((((((((((((((((....((((((((((.............)))))))))).)))))))))))))))) ( -42.32) >DroPse_CAF1 1414 113 - 1 GAAGCUCAUUAGAAGAGGAAACACAAUUUUCAGAGGACUUCAGGGGAACAUCCCAUCGAGGAGGACCCCU-CCAGUGGGUGUGUC-UGGCUGGAGGAGGCCGCCUUUGGUUGCGC ...((...........(....).......(((((((.((((..(((.....)))...)))).((.(((((-(((((.((.....)-).)))))))).)))).)))))))..)).. ( -43.90) >DroSec_CAF1 2541 112 - 1 CAUUCUCUCAAGAAU-UGAAACGCAGUUG--ACAGGAGUUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGUAGUUUUUGGAGUAAGUGGUCGUGGGUUCAUC ....((((....(((-((.....))))).--...)))).......((((((((((((((((.......(((((((.............)))))))....)))))))))))))))) ( -40.32) >DroSim_CAF1 2557 112 - 1 CAUUCUCUCAAGAAU-UGAAACGCAGUUG--ACAGGAGAUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGUAGUUUUUGGAGUAACUGGUCGUGGGUUCAUC ...(((((....(((-((.....))))).--...)))))......((((((((((((((((..(((..(((((((.............))))))).))))))))))))))))))) ( -43.32) >DroEre_CAF1 2517 112 - 1 CAUUCUCUCAAGAAU-UGAAACGCAGUUG--ACAGGAGUUCUAAGGAUGAGCCCGCGACCAAAGCUUCCUCCUAGUGGGUGUGUAAAUUUUGGAGUAGCUCGUCGUGGGUUCAUC ....((((....(((-((.....))))).--...)))).......((((((((((((((...((((..((((....((((......)))).)))).)))).)))))))))))))) ( -38.20) >DroYak_CAF1 2545 112 - 1 CAUUCUCUUAAGUAU-UGAAACGCUGUUG--ACAGGAGUUCCAAGGAUGAGCCCGCGACCAAAGCUACCUCCUGGUGGGUGUGUAGUUUUUGGAGUAGCUGAUCGUGGGUUCAUC ..............(-((.(((.(((...--.)))..))).))).(((((((((((((....((((((.(((...................)))))))))..))))))))))))) ( -38.31) >consensus CAUUCUCUCAAGAAU_UGAAACGCAGUUG__ACAGGAGUUCCAAGGAUGAGCCCGCGACCAAAGUUUCCUCCAAGUGGGUGUGUAGUUUUUGGAGUAACUGGUCGUGGGUUCAUC ..................................((....))...((((((((((((((((.......((((...................))))....)))))))))))))))) (-24.44 = -24.89 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:14 2006