
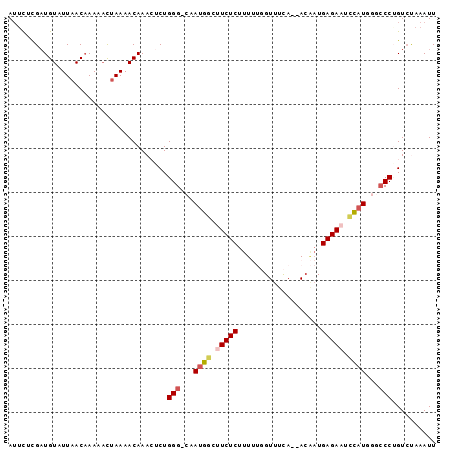
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,628,347 – 11,628,475 |
| Length | 128 |
| Max. P | 0.774836 |

| Location | 11,628,347 – 11,628,443 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -10.67 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11628347 96 + 20766785 AUUCUCGUUGUAUUAACAAAACUAAAAACAAACUCUGGG-CAAUGACUUCUCUUUUUGGUUUCAUUAGAAUGAGAACUCAUUGGCCCUGUCUAAAUU .......((((....)))).................(((-((((((.(((((.(((((((...))))))).))))).))))).)))).......... ( -21.10) >DroSec_CAF1 896 94 + 1 AUUCUCGAUGUAUUAACAAAAACUAAAACAAACUCUGGG-CAAUGGCCUCUCUUUUUGGUUUCA--ACAAUGAGAAUCCAUGGGCCCAGUCCAAAUU ........(((.(((........))).)))....(((((-(.((((..((((...(((......--.))).))))..))))..))))))........ ( -23.10) >DroSim_CAF1 899 94 + 1 AUUCUCGAUGUAUUAACAAAAACUAAAACAAACUCUGGG-CAAUGGCCUCUCUUUUUGGUUUCA--ACAAUGAGAAUCCAUGGGCCCAGUCCAAAUU ........(((.(((........))).)))....(((((-(.((((..((((...(((......--.))).))))..))))..))))))........ ( -23.10) >DroEre_CAF1 879 90 + 1 AAAUCUGAUGUAUUAACAAA-ACUAAAACAAAGGCUGGG-CAAUGGCUUCUCUUU---GUUUUA--ACGAUGAGAAUCCAUGAAGCCUGUCUAAAUU ....................-..........((((.(((-(.((((.(((((.((---((....--)))).))))).))))...))))))))..... ( -22.40) >DroYak_CAF1 895 91 + 1 AUUCUCGAUGUAUUAACCAA-ACUAAAACAAAGUCUGGGACAAUGACUUCUCUUU---GUUUCA--ACCCUGAGAAUCUUUGAACCCUGUAUAAAUU (((((((..((.....(((.-(((.......))).)))(((((.((....)).))---)))...--))..))))))).................... ( -12.90) >consensus AUUCUCGAUGUAUUAACAAAAACUAAAACAAACUCUGGG_CAAUGGCUUCUCUUUUUGGUUUCA__ACAAUGAGAAUCCAUGGGCCCUGUCUAAAUU ....................................(((...((((.(((((...................))))).))))...))).......... (-10.67 = -11.27 + 0.60)


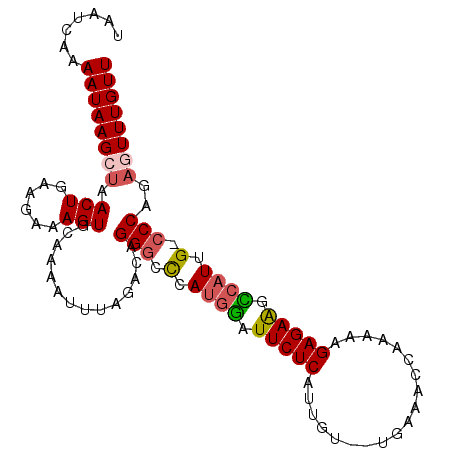
| Location | 11,628,373 – 11,628,475 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -14.91 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11628373 102 - 20766785 UAAUCAAAAUAAGCUAACUGAAGAAAGUGCAAAAUUUAGACAGGGCCAAUGAGUUCUCAUUCUAAUGAAACCAAAAAGAGAAGUCAUUG-CCCAGAGUUUGUU .......((((((((..(((((............)))))...((((.(((((.(((((...................))))).))))))-)))..)))))))) ( -25.71) >DroSec_CAF1 922 100 - 1 UAAUCAAAAUAAGCUAACUGAAGAAAGUGCAAAAUUUGGACUGGGCCCAUGGAUUCUCAUUGU--UGAAACCAAAAAGAGAGGCCAUUG-CCCAGAGUUUGUU .......((((((((.(((......)))............((((((..((((.(((((....(--((....)))...))))).)))).)-))))))))))))) ( -28.30) >DroSim_CAF1 925 100 - 1 UAAUCAAAAUAAGCUAACUGAAGAAAGUGCAAAAUUUGGACUGGGCCCAUGGAUUCUCAUUGU--UGAAACCAAAAAGAGAGGCCAUUG-CCCAGAGUUUGUU .......((((((((.(((......)))............((((((..((((.(((((....(--((....)))...))))).)))).)-))))))))))))) ( -28.30) >DroEre_CAF1 904 97 - 1 UAAUCAAAAUAAGCUAACUGAAGAAAGUGCAAAAUUUAGACAGGCUUCAUGGAUUCUCAUCGU--UAAAAC---AAAGAGAAGCCAUUG-CCCAGCCUUUGUU ............(((..(((((............)))))...(((...((((.(((((...((--....))---...))))).)))).)-)).)))....... ( -16.80) >DroYak_CAF1 920 98 - 1 UAAUCAAAAUAAGCUAACUGAAGAAAGUGCAAAAUUUAUACAGGGUUCAAAGAUUCUCAGGGU--UGAAAC---AAAGAGAAGUCAUUGUCCCAGACUUUGUU ....((((....((..(((......)))))............(((..(((.(((((((...((--....))---...))).)))).))).)))....)))).. ( -15.40) >consensus UAAUCAAAAUAAGCUAACUGAAGAAAGUGCAAAAUUUAGACAGGGCCCAUGGAUUCUCAUUGU__UGAAACCAAAAAGAGAAGCCAUUG_CCCAGAGUUUGUU .......((((((((.(((......)))..............(((.(.((((.(((((...................))))).)))).).)))..)))))))) (-14.91 = -15.23 + 0.32)

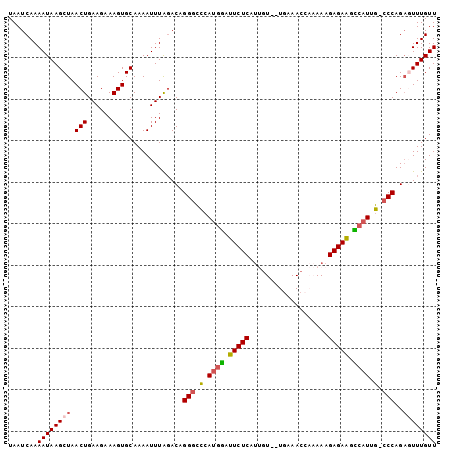
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:07 2006