| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,605,604 – 11,605,700 |
| Length | 96 |
| Max. P | 0.764577 |

| Location | 11,605,604 – 11,605,700 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.40 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
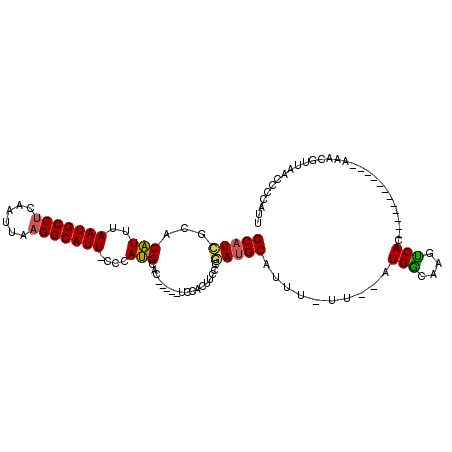
>2R_DroMel_CAF1 11605604 96 - 20766785 GCAUCGCACAUUUAAUGCCGCAAUUAAGGCAUU-CCCAUGCACUUCACUCCACUUCCCUAUGCAUUU-UU--AUGACAAGUCAC------------AAACAUUAACCCCAUU .....(((.....((((((........))))))-....)))...................(((((..-..--))).))......------------................ ( -11.30) >DroSec_CAF1 3679 93 - 1 GCAUCGCACAUUUAAUGCCUCAAUUAAGGCAUU-CCCAUGCAC-----UCCACUUCCCGAUGCGUUC-UUAAAUGGCAAGUCAC------------AAACGUUAACCCCAUU .....((.((((((((((((......)))))..-...(((((.-----((........)))))))..-))))))))).......------------................ ( -17.00) >DroSim_CAF1 3710 98 - 1 GCAUCGCACAUUUAAUGCCUCAAUUAAGGCAUU-CCCAUGCACUUUACUCCACUUCCCGAUGCGUUU-UUUAAUGGCAAGUCAC------------AAACGUUAACCCCAUU ((((((((.....(((((((......)))))))-....))).................)))))....-.((((((.........------------...))))))....... ( -15.92) >DroEre_CAF1 3601 91 - 1 GCAUCCCACAUUUAAUGCCUCAAUUAAGGCAUU-CCCAUGCAC-----UCCACUUCCCGAUGCAUUU-UU--AUGGCAAGUCAC------------AAACGUUAACCCCAUU ((...........(((((((......)))))))-...(((((.-----((........)))))))..-..--...)).......------------................ ( -13.30) >DroYak_CAF1 4390 91 - 1 GCAUCCCACAUUUAAUGCCUCAAUUAAGGCAUU-CCCAUGCAC-----UUCACUUCGCGAUGCAUUU-UU--AUGGCAAGUCAC------------AAACGUUAACCCCAUU (((((........(((((((......)))))))-.....((..-----........)))))))....-..--((((......((------------....)).....)))). ( -15.40) >DroPer_CAF1 5941 112 - 1 GCUUGUCUCGUUUAAUGCCUCAAUUAAGGCAUUUCACACGCAGUUUUUAUGCGUUUUCCAUGCACAUUUUAAAUGGCUUUCCACUGCCCCGAGCUAAAACGUUAACCCCAUU (((((....((..(((((((......)))))))..))..(((((.....(((((.....)))))..........((....)))))))..))))).................. ( -24.90) >consensus GCAUCGCACAUUUAAUGCCUCAAUUAAGGCAUU_CCCAUGCAC_____UCCACUUCCCGAUGCAUUU_UU__AUGGCAAGUCAC____________AAACGUUAACCCCAUU (((((...(((..(((((((......)))))))....)))..................)))))..........(((....)))............................. (-10.68 = -10.40 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:53 2006