
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
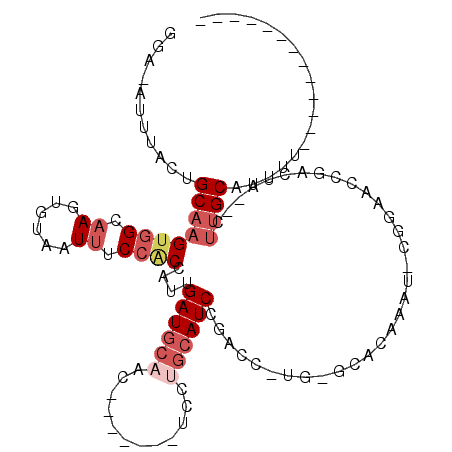
| Location | 2,283,593 – 2,283,686 |
| Length | 93 |
| Max. P | 0.957765 |

| Location | 2,283,593 – 2,283,686 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.01 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -8.13 |
| Energy contribution | -9.47 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
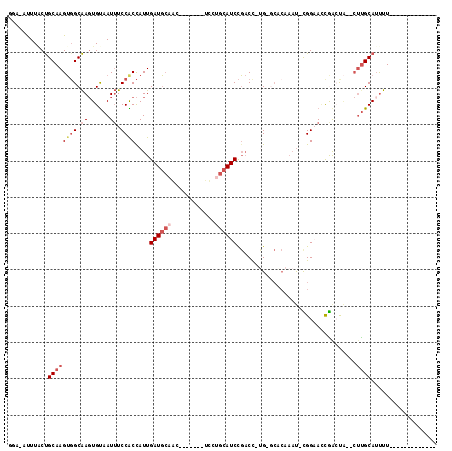
>2R_DroMel_CAF1 2283593 93 + 20766785 GGA-AUUUACUGCAAGUGGCAAGUGUAAUUUCCACCAUUGAUGCAAC-------UUCUGCAUCCGACC-UGAGCACAAAU-CGGAACCGGCUA--CUCGCAUUUU------------- ...-......(((.((((((..((((....((.......((((((..-------...)))))).....-.))))))....-((....))))))--)).)))....------------- ( -22.82) >DroSec_CAF1 49711 93 + 1 GGA-AUUUACUGCAAGUGGCAAGUGUAAUUUCCACCAUUGAUGCAAC-------UUCUGCAUCCGACC-UGUGCACAAAU-CGGAAUCGACCA--CUUGCAUUUU------------- ...-......(((((((((...(((.......)))..((((((((..-------...))).(((((..-((....))..)-))))))))))))--))))))....------------- ( -26.70) >DroSim_CAF1 50396 91 + 1 -----UUUACUGCAAGUGGCAAGUGUAAUUUCCACCAUUGAUGCAAC-------UUCUGCAUCCGACCNUGGGCACAAAUNCGGAACCGACCA--CUUGCAUUUU------------- -----.....(((((((((...((((.....(((.....((((((..-------...))))))......))))))).....((....)).)))--))))))....------------- ( -28.00) >DroEre_CAF1 50150 99 + 1 CGG-AUUUUCUGCAAGUGGCAAGUGUAAUUUCCGCCAUUGAUGCAGUCAAUGCAUCCUGCAUCCGACU-GG-GCUCAAAU-UGGCACCAAAUC--CUUGCAUUUU------------- .((-((((..(((.((((((.............))))))(((.(((((.((((.....))))..))))-).-).))....-..)))..)))))--).........------------- ( -28.02) >DroYak_CAF1 45485 93 + 1 GGA-ACUUGCUGCAAGUUGCAACUGUAAUUUCCGCUAUUGAUGCCAU-------UCCUGCAUCUGACU-UGGCCUCAAAU-UGGAACCGACUA--CUUGCAUUUU------------- ..(-(((((...))))))((((..((...(((((...((((.((((.-------((........))..-)))).))))..-)))))...))..--.)))).....------------- ( -21.00) >DroPer_CAF1 51087 105 + 1 CCACAUUUACCGCAAGAGGCGAGUGCAAUAUCCGCCAUUGAUUGCGCCA---CAACUAACAUCCUCCU-GG--------C-UGGCAGUAAAUGAGGCUGCAGCCUGGAAACCAAACCC ((.(((((((.((....((((....((((.......))))....)))).---.......((.((....-))--------.-)))).)))))))((((....))))))........... ( -29.10) >consensus GGA_AUUUACUGCAAGUGGCAAGUGUAAUUUCCACCAUUGAUGCAAC_______UCCUGCAUCCGACC_UG_GCACAAAU_CGGAACCGACUA__CUUGCAUUUU_____________ ...........((((((((.((......)).))))....((((((............)))))).................................)))).................. ( -8.13 = -9.47 + 1.33)

| Location | 2,283,593 – 2,283,686 |
|---|---|
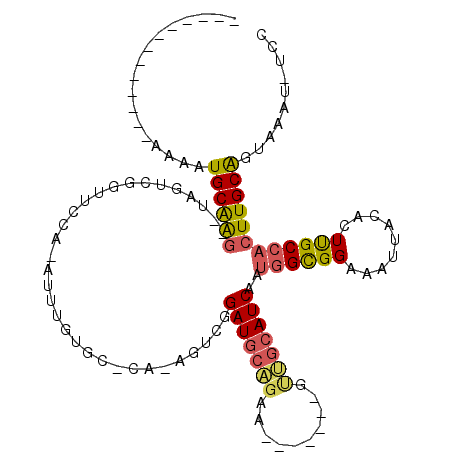
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.01 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
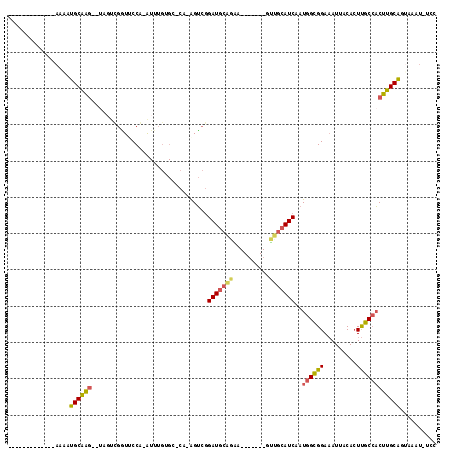
>2R_DroMel_CAF1 2283593 93 - 20766785 -------------AAAAUGCGAG--UAGCCGGUUCCG-AUUUGUGCUCA-GGUCGGAUGCAGAA-------GUUGCAUCAAUGGUGGAAAUUACACUUGCCACUUGCAGUAAAU-UCC -------------....((((((--(.((.(((((((-(((((....))-)))).(((((((..-------.)))))))......)))......))).)).)))))))......-... ( -28.10) >DroSec_CAF1 49711 93 - 1 -------------AAAAUGCAAG--UGGUCGAUUCCG-AUUUGUGCACA-GGUCGGAUGCAGAA-------GUUGCAUCAAUGGUGGAAAUUACACUUGCCACUUGCAGUAAAU-UCC -------------....((((((--((((.((.((((-(((((....))-)))))))(((((..-------.)))))))...((((.......)))).))))))))))......-... ( -33.50) >DroSim_CAF1 50396 91 - 1 -------------AAAAUGCAAG--UGGUCGGUUCCGNAUUUGUGCCCANGGUCGGAUGCAGAA-------GUUGCAUCAAUGGUGGAAAUUACACUUGCCACUUGCAGUAAA----- -------------....((((((--((((.(((.....((((.((((........(((((((..-------.)))))))...)))).))))...))).)))))))))).....----- ( -30.20) >DroEre_CAF1 50150 99 - 1 -------------AAAAUGCAAG--GAUUUGGUGCCA-AUUUGAGC-CC-AGUCGGAUGCAGGAUGCAUUGACUGCAUCAAUGGCGGAAAUUACACUUGCCACUUGCAGAAAAU-CCG -------------.........(--(((((..(((..-..((((..-.(-(((((.((((.....))))))))))..))))((((((.........))))))...)))..))))-)). ( -30.60) >DroYak_CAF1 45485 93 - 1 -------------AAAAUGCAAG--UAGUCGGUUCCA-AUUUGAGGCCA-AGUCAGAUGCAGGA-------AUGGCAUCAAUAGCGGAAAUUACAGUUGCAACUUGCAGCAAGU-UCC -------------.........(--((((...((((.-..((((.((((-..((.......)).-------.)))).))))....))))))))).(((((.....)))))....-... ( -22.50) >DroPer_CAF1 51087 105 - 1 GGGUUUGGUUUCCAGGCUGCAGCCUCAUUUACUGCCA-G--------CC-AGGAGGAUGUUAGUUG---UGGCGCAAUCAAUGGCGGAUAUUGCACUCGCCUCUUGCGGUAAAUGUGG .(((((((...)))))))....((.((((((((((..-(--------((-(.((.........)).---)))).........(((((.........)))))....)))))))))).)) ( -33.50) >consensus _____________AAAAUGCAAG__UAGUCGGUUCCA_AUUUGUGC_CA_AGUCGGAUGCAGAA_______GUUGCAUCAAUGGCGGAAAUUACACUUGCCACUUGCAGUAAAU_UCC .................((((((................................(((((((..........)))))))..((((((.........)))))))))))).......... (-15.09 = -15.12 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:02 2006