
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,592,236 – 11,592,429 |
| Length | 193 |
| Max. P | 0.933291 |

| Location | 11,592,236 – 11,592,356 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11592236 120 - 20766785 AGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCCGCGACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUUGUUGUGGGAGUACUCAGCUCCUGCCUGCCUGCCACUCAUC ((((.(((((...............(((((.....)))))((((....))))....))))).))))....((.((.((..((.(..(((((.....)))))..)..))..)).))))... ( -39.40) >DroSec_CAF1 25789 118 - 1 AGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCACC--GACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUUGUUGUGGGAGUACUCAGCUCCUGCCUGCCUGCCACUCAUC ((((.(((((.....((((......)))).........--((((....))))....))))).))))....((.((.((..((.(..(((((.....)))))..)..))..)).))))... ( -35.30) >DroSim_CAF1 29072 118 - 1 AGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCC--GACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUUGUUGUGGGAGUACUCAGCUCCUGCCUGCCUGCCACUCAUC ((((.(((((.....((((......)))).........--((((....))))....))))).))))....((.((.((..((.(..(((((.....)))))..)..))..)).))))... ( -35.30) >consensus AGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCC__GACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUUGUUGUGGGAGUACUCAGCUCCUGCCUGCCUGCCACUCAUC ((((.(((((.....((((......))))...........((((....))))....))))).))))....((.((.((..((.(..(((((.....)))))..)..))..)).))))... (-35.10 = -35.10 + 0.00)



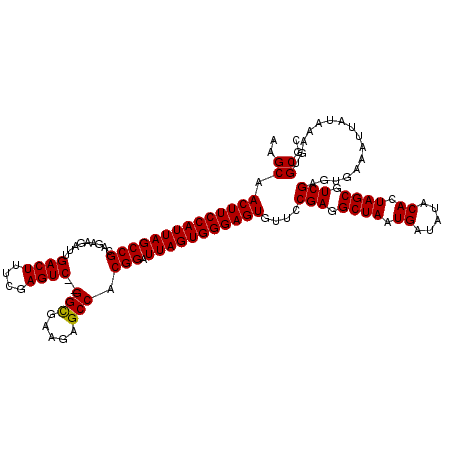
| Location | 11,592,276 – 11,592,396 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -36.69 |
| Energy contribution | -36.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11592276 120 + 20766785 AAGCAACUUCCAUUAGCCGCAGAAGAUUGACUUUCGAGUCGCGGCGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAGUGAAAUUAUAAAGUGCGC ..(((((((((((((((((.........((((....))))..(((.....))).))).)))))))))))....(((.((((.((.....)).)))).)))...............))).. ( -39.40) >DroSec_CAF1 25829 118 + 1 AAGCAACUUCCAUUAGCCGCAGAAGAUUGACUUUCGAGUC--GGUGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAGUGAAAUUAUAAAGAGCGC ..((.((((((((((((((.........((((....))))--(((.....))).))).)))))))))))....(((.((((.((.....)).)))).)))................)).. ( -36.40) >DroSim_CAF1 29112 118 + 1 AAGCAACUUCCAUUAGCCGCAGAAGAUUGACUUUCGAGUC--GGCGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAAUGAAAUUAUAAAGUGCGC ..(((((((((((((((((.........((((....))))--(((.....))).))).)))))))))))....(((.((((.((.....)).)))).)))...............))).. ( -39.40) >consensus AAGCAACUUCCAUUAGCCGCAGAAGAUUGACUUUCGAGUC__GGCGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAGUGAAAUUAUAAAGUGCGC ..((.((((((((((((((.........((((....))))..(((.....))).))).)))))))))))....(((.((((.((.....)).)))).)))................)).. (-36.69 = -36.47 + -0.22)

| Location | 11,592,276 – 11,592,396 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -30.45 |
| Energy contribution | -30.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11592276 120 - 20766785 GCGCACUUUAUAAUUUCACUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCCGCGACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUU (((.(((((.......((((.......)))).....((((((((.(((((...............(((((.....)))))((((....))))....))))).)))))))))))))))).. ( -36.80) >DroSec_CAF1 25829 118 - 1 GCGCUCUUUAUAAUUUCACUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCACC--GACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUU (((..((((.......((((.......)))).....((((((((.(((((.....((((......)))).........--((((....))))....))))).)))))))))))).))).. ( -30.90) >DroSim_CAF1 29112 118 - 1 GCGCACUUUAUAAUUUCAUUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCC--GACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUU ..(((.............(((((.(((((((.....))))))).)))))..(((.(((.((..(((((((.....)))--((((....))))........)))).)).))).)))))).. ( -33.10) >consensus GCGCACUUUAUAAUUUCACUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCC__GACUCGAAAGUCAAUCUUCUGCGGCUAAUGGAAGUUGCUU (((.(((((.......((((.......)))).....((((((((.(((((.....((((......))))...........((((....))))....))))).)))))))))))))))).. (-30.45 = -30.57 + 0.11)

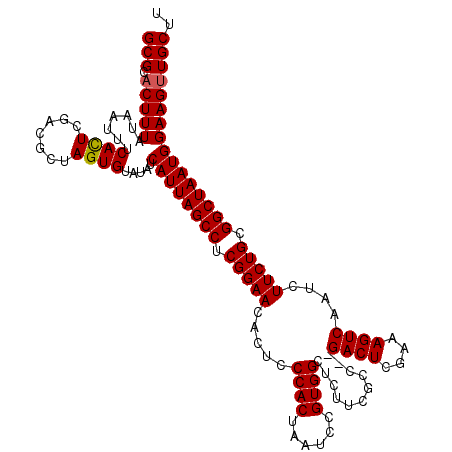
| Location | 11,592,316 – 11,592,429 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -30.05 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11592316 113 + 20766785 GCGGCGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAGUGAAAUUAUAAAGUGCGCAGCAGCCACUACAAUUAAUUUCGGAGCA--U-----UGCA ...((......((((......)))).((((((((((.((((.((.....)).)))).))....(((((((...((((.((....)))))).....)))))))))))))--)-----))). ( -37.50) >DroSec_CAF1 25869 111 + 1 --GGUGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAGUGAAAUUAUAAAGAGCGCAGCAGCCACUACAAUUAAUUUCGGAGCA--U-----UGCA --.........((((......)))).((((((((((.((((.((.....)).)))).))....(((((((.....((.((....))..)).....)))))))))))))--)-----)... ( -31.20) >DroSim_CAF1 29152 117 + 1 --GGCGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAAUGAAAUUAUAAAGUGCGCAGCAGCCACUACAAUUAAU-UCGGAGCGAAUCCCACCACA --(((.....)))........((((((.((((((((.((((.((.....)).)))).))((((..((((....((((.((....))))))..)))).))-))))))))..)))))).... ( -39.40) >consensus __GGCGAAGAGCCACGGAUUAGUGGGAGUGUUCCGAGGCUAAUGAUAUACACUAGCGUCGAGUGAAAUUAUAAAGUGCGCAGCAGCCACUACAAUUAAUUUCGGAGCA__U_____UGCA ...(((.....((((......))))...((((((((.((((.((.....)).)))).))....(((((((...((((.((....)))))).....)))))))))))))........))). (-30.05 = -30.50 + 0.45)

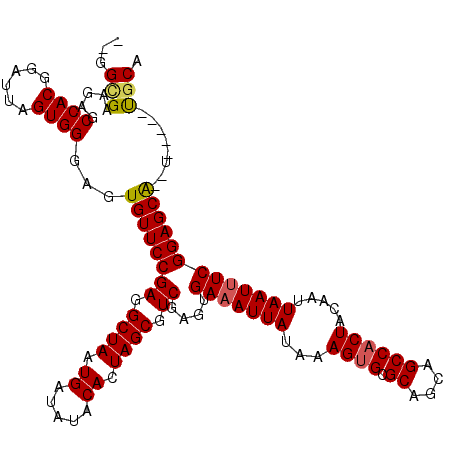
| Location | 11,592,316 – 11,592,429 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -28.53 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11592316 113 - 20766785 UGCA-----A--UGCUCCGAAAUUAAUUGUAGUGGCUGCUGCGCACUUUAUAAUUUCACUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCCGC ....-----.--((.((((((((((.....((((((....)).))))...)))))))....((.(((((((.....))))))).))))).)).............(((((.....))))) ( -32.40) >DroSec_CAF1 25869 111 - 1 UGCA-----A--UGCUCCGAAAUUAAUUGUAGUGGCUGCUGCGCUCUUUAUAAUUUCACUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCACC-- ....-----.--((.((((((((((..((((((....)))))).......)))))))....((.(((((((.....))))))).))))).))...((((......)))).........-- ( -26.60) >DroSim_CAF1 29152 117 - 1 UGUGGUGGGAUUCGCUCCGA-AUUAAUUGUAGUGGCUGCUGCGCACUUUAUAAUUUCAUUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCC-- ..((((((((...(.(((((-((.((((((((((((....)).))))..))))))..))))((.(((((((.....))))))).))))).)..))))))))......(((.....)))-- ( -38.90) >consensus UGCA_____A__UGCUCCGAAAUUAAUUGUAGUGGCUGCUGCGCACUUUAUAAUUUCACUCGACGCUAGUGUAUAUCAUUAGCCUCGGAACACUCCCACUAAUCCGUGGCUCUUCGCC__ .((((......))))((((((((((.....((((((....)).))))...)))))))....((.(((((((.....))))))).)))))......((((......))))........... (-28.53 = -28.70 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:45 2006