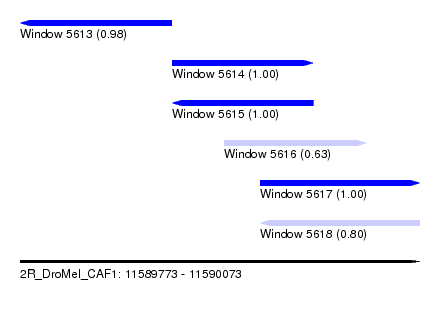
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,589,773 – 11,590,073 |
| Length | 300 |
| Max. P | 0.999634 |

| Location | 11,589,773 – 11,589,887 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.25 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -37.00 |
| Energy contribution | -37.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11589773 114 - 20766785 GAAA------UGGAAAAGUACGGACACACCGAGAGAGAUUUGCAUUCGUAACUCGAUUUGCGGUGUGAGCUCUCAUCGCCACUCUCAGUCGGUUGCCCAGGAUCCUGUGGCAUGCAAAAU ....------.......(((.......((((((((((....(((.(((.....)))..)))(((((((....))).)))).)))))..)))))(((((((....))).)))))))..... ( -37.00) >DroSec_CAF1 23346 114 - 1 GAAA------UGGAAAAGUACGGACACACCGAGAGAGAUUUGCAUUCGUAACUCGAUUUGCGGUGUGAGCUCUCAUCGCCACUCUCAGUCGGUUGCCCAGGAUCCUGUGGCAUGCAAAAU ....------.......(((.......((((((((((....(((.(((.....)))..)))(((((((....))).)))).)))))..)))))(((((((....))).)))))))..... ( -37.00) >DroSim_CAF1 26624 120 - 1 GAAAUGGAAAUGGAAAAGUACGGACACACCGAGAGAGAUUUGCAUUCGUAACUCGAUUUGCGGUGUGAGCUCUCAUCGCCACUCUCAGUCGGUUGCCCAGGAUCCUGUGGCAUGCAAAAU .................(((.......((((((((((....(((.(((.....)))..)))(((((((....))).)))).)))))..)))))(((((((....))).)))))))..... ( -37.00) >DroEre_CAF1 24012 114 - 1 GAAA------UGGAAAAGUACGGACACACCGAGAGAGAUUUGCAUUCGUAACUCGAUUUGCGGUGUGAGCUCUCAUCGCCACUCUCAGUCGGUUGCCCAGGAUCCUGUGGCAUGCAAAAU ....------.......(((.......((((((((((....(((.(((.....)))..)))(((((((....))).)))).)))))..)))))(((((((....))).)))))))..... ( -37.00) >DroYak_CAF1 24138 112 - 1 --AA------UGGAAAAGUACGGACACACCGAGAGAGAUUUGCAUUCGUAACUCGAUUUGCGGUGUGAGCUCUCAUCGCCACUCUCAGUCGGUUGCCCAGGAUCCUGUGGCAUGCAAAAU --..------.......(((.......((((((((((....(((.(((.....)))..)))(((((((....))).)))).)))))..)))))(((((((....))).)))))))..... ( -37.00) >consensus GAAA______UGGAAAAGUACGGACACACCGAGAGAGAUUUGCAUUCGUAACUCGAUUUGCGGUGUGAGCUCUCAUCGCCACUCUCAGUCGGUUGCCCAGGAUCCUGUGGCAUGCAAAAU .................(((.......((((((((((....(((.(((.....)))..)))(((((((....))).)))).)))))..)))))(((((((....))).)))))))..... (-37.00 = -37.00 + 0.00)



| Location | 11,589,887 – 11,589,993 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -24.84 |
| Energy contribution | -27.22 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11589887 106 + 20766785 CACGCCAUUCUUUUUUUUUUUUUUUUGGCCAAGCCG-UUUGGAGC-------UAGAUGAACUGGAGAAGUUGGU-CGGUUGGCUUGGCCGGCGUGUCCAGAAAUGGACACUGCGA ..(((...................((((((((((((-......((-------(.(((.((((.....)))).))-)))))))))))))))).(((((((....))))))).))). ( -44.20) >DroSec_CAF1 23460 91 + 1 CACGCCAUUCUAUUUU----CUUUUUGGCCAAGCCGGUUUGCAGC-------UAGAUGAACUGGAGAAG-------------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGA ...((((.........----.....))))...(((((((.((..(-------(((.....))))....)-------------)..)))))))(((((((....)))))))..... ( -35.04) >DroSim_CAF1 26744 91 + 1 CACGCCAUUCCAUUUU----CUUUUUGGCCAAGCCGGUUUGCAGC-------UAGAUGAACUGGAGAAG-------------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGA ...((((.........----.....))))...(((((((.((..(-------(((.....))))....)-------------)..)))))))(((((((....)))))))..... ( -35.04) >DroEre_CAF1 24126 100 + 1 CACGCCAU-----UUU----CUUUUUGGCCAAGCCGGA-UGAAGCCGGCACACAGACGAACUGGAGAAGCUGGCU-----GGCUUGGUCGGCGUGUCCAGAAGUGGACACUGCGA ..(((...-----...----....(((((((((((...-...(((((((...(((.....))).....)))))))-----))))))))))).(((((((....))))))).))). ( -43.50) >consensus CACGCCAUUCUAUUUU____CUUUUUGGCCAAGCCGGUUUGCAGC_______UAGAUGAACUGGAGAAG_____________CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGA ..(((...................(((((((((((((((((...............))))))))..................))))))))).(((((((....))))))).))). (-24.84 = -27.22 + 2.38)

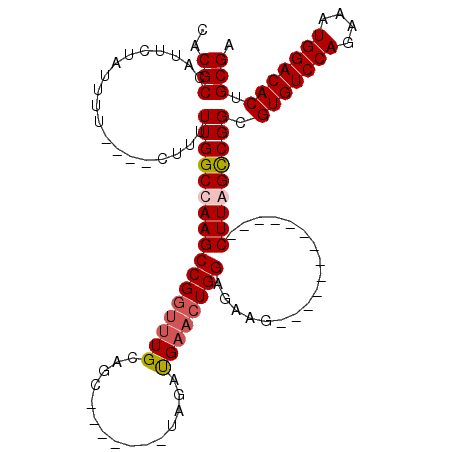
| Location | 11,589,887 – 11,589,993 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.36 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11589887 106 - 20766785 UCGCAGUGUCCAUUUCUGGACACGCCGGCCAAGCCAACCG-ACCAACUUCUCCAGUUCAUCUA-------GCUCCAAA-CGGCUUGGCCAAAAAAAAAAAAAAAAGAAUGGCGUG .(((.(((((((....)))))))...(((((((((....(-(......))...((((.....)-------))).....-.))))))))).....................))).. ( -33.90) >DroSec_CAF1 23460 91 - 1 UCGCAGUGUCCAUUUCUGGACACGCCGGCUAAG-------------CUUCUCCAGUUCAUCUA-------GCUGCAAACCGGCUUGGCCAAAAAG----AAAAUAGAAUGGCGUG .(((.(((((((....)))))))...(((((((-------------((....(((((.....)-------))))......)))))))))......----...........))).. ( -32.30) >DroSim_CAF1 26744 91 - 1 UCGCAGUGUCCAUUUCUGGACACGCCGGCUAAG-------------CUUCUCCAGUUCAUCUA-------GCUGCAAACCGGCUUGGCCAAAAAG----AAAAUGGAAUGGCGUG .(((.(((((((....)))))))...(((((((-------------((....(((((.....)-------))))......)))))))))......----...........))).. ( -32.30) >DroEre_CAF1 24126 100 - 1 UCGCAGUGUCCACUUCUGGACACGCCGACCAAGCC-----AGCCAGCUUCUCCAGUUCGUCUGUGUGCCGGCUUCA-UCCGGCUUGGCCAAAAAG----AAA-----AUGGCGUG .(((.(((((((....)))))))((((.(((((((-----((((.((....((((.....))).).)).))))...-...))))))).(.....)----...-----.))))))) ( -36.20) >consensus UCGCAGUGUCCAUUUCUGGACACGCCGGCCAAG_____________CUUCUCCAGUUCAUCUA_______GCUGCAAACCGGCUUGGCCAAAAAG____AAAAUAGAAUGGCGUG .(((.(((((((....)))))))...(((((((((.............................................))))))))).....................))).. (-23.35 = -24.36 + 1.00)

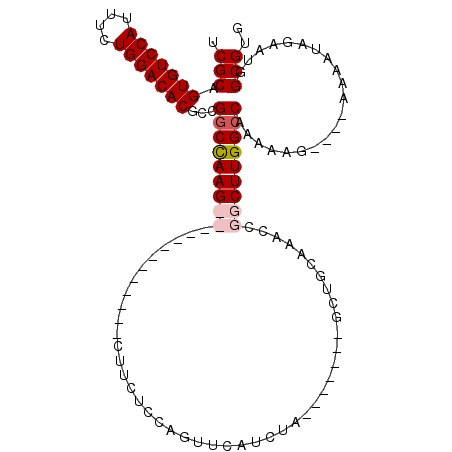
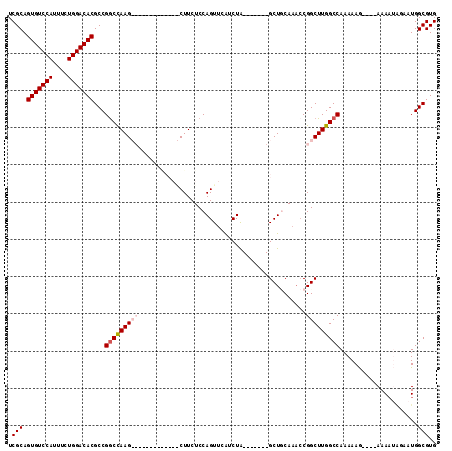
| Location | 11,589,926 – 11,590,033 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -24.66 |
| Energy contribution | -26.82 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11589926 107 + 20766785 GGAGC--------UAGAUGAACUGGAGAAGUUGGU-----CGGUUGGCUUGGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUU ((.((--------..(((((((((.....((((((-----(((.....)))))))))(((((((....)))))))(((.((.....)).)))....)).)).)))))..))))....... ( -37.80) >DroSec_CAF1 23496 95 + 1 GCAGC--------UAGAUGAACUGGAGAAG-----------------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUU (((((--------..(.(((.((.....))-----------------.))).)..))(((((((....))))))))))....(((((.(((....((....))......))))))))... ( -27.80) >DroSim_CAF1 26780 95 + 1 GCAGC--------UAGAUGAACUGGAGAAG-----------------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUU (((((--------..(.(((.((.....))-----------------.))).)..))(((((((....))))))))))....(((((.(((....((....))......))))))))... ( -27.80) >DroEre_CAF1 24156 110 + 1 GAAGCCG-GCACACAGACGAACUGGAGAAGCUGGCU---------GGCUUGGUCGGCGUGUCCAGAAGUGGACACUGCGACAAUUUGAGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUU ..(((((-((...(((.....))).....)))))))---------(((...((((..(((((((....)))))))..))))....(((.(((......))).)))....)))........ ( -36.50) >DroYak_CAF1 24270 120 + 1 GAAGCUACAUACAUAGAUGUACUGGAGCAGCUGGCUAGCUUGACUGGCUUAGCUGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGAGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUU ..((.(((((......)))))))(..(((((..(((((((.....))).))))..))(((((((....))))))))))..).(((((.(((....((....))......))))))))... ( -42.90) >consensus GAAGC________UAGAUGAACUGGAGAAG_UGG___________GGCUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUU (..((................((((..(((((((.........)))))))..)))).(((((((....))))))).))..).(((((.(((....((....))......))))))))... (-24.66 = -26.82 + 2.16)


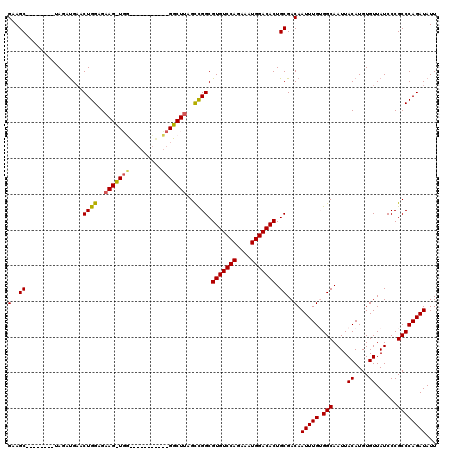
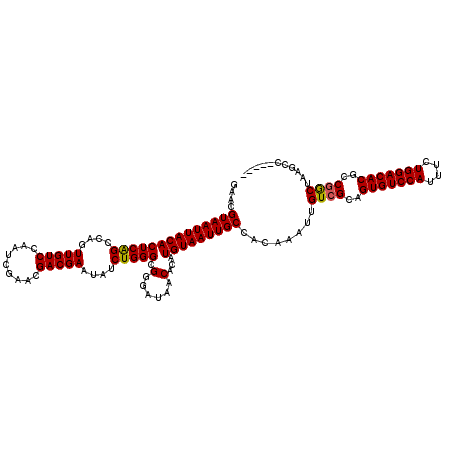
| Location | 11,589,953 – 11,590,073 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -37.44 |
| Energy contribution | -37.12 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11589953 120 + 20766785 CGGUUGGCUUGGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUUCGUCGUUCGAUUGGACAACUGGCUGAGUGUAAUUACGUUC .((((((((..(((((.(((((((....))))))).((((..(((((.(((....((....))......))))))))..)))).((((....))))..)))))..))).)))))...... ( -41.20) >DroSec_CAF1 23518 113 + 1 -------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUUCGUCGUUCGAUUGGACAACUGGCUGAGUGUAAUUACGUUC -------(((((((((.(((((((....))))))).((((..(((((.(((....((....))......))))))))..)))).((((....))))..)))))))))............. ( -39.20) >DroSim_CAF1 26802 113 + 1 -------CUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUUCGUCGUUCGAUUGGACAACUGGCUGAGUGUAAUUACGUUC -------(((((((((.(((((((....))))))).((((..(((((.(((....((....))......))))))))..)))).((((....))))..)))))))))............. ( -39.20) >DroEre_CAF1 24191 115 + 1 -----GGCUUGGUCGGCGUGUCCAGAAGUGGACACUGCGACAAUUUGAGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUUCGUCGUUCGAUUGGACAACUGGCCGAGUGUAAUUACUUUC -----.((((((((((..(((((((...(((((...((((..(((((.(((....((....))......))))))))..)))).))))).))))))).))))))))))............ ( -39.40) >DroYak_CAF1 24310 120 + 1 UGACUGGCUUAGCUGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGAGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUUCGUCGUUCGAUUGGACAACUGGCUGAGUGUAAUUACUUUC ......(((((((..(.(((((((....))))))).((((..(((((.(((....((....))......))))))))..)))).((((....))))..)..)))))))............ ( -39.10) >consensus _____GGCUUAGCCGGCGUGUCCAGAAAUGGACACUGCGACAAUUUGUGGCAAUUACAUGUGUUAUCCCGCCCAGAUAUUCGUCGUUCGAUUGGACAACUGGCUGAGUGUAAUUACGUUC ......((((((((((..(((((((...(((((...((((..(((((.(((....((....))......))))))))..)))).))))).))))))).))))))))))............ (-37.44 = -37.12 + -0.32)

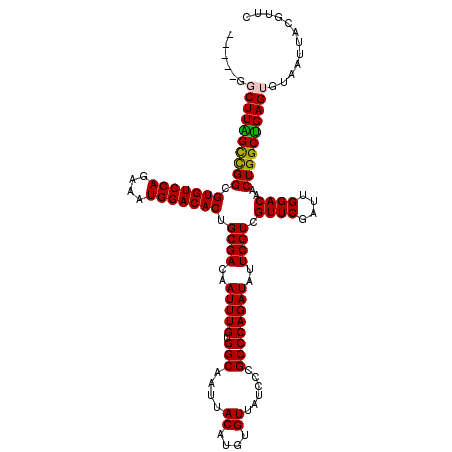
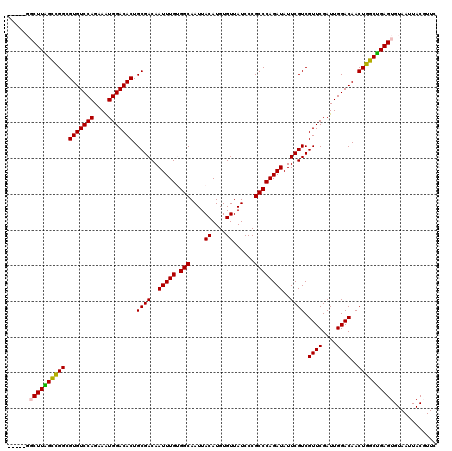
| Location | 11,589,953 – 11,590,073 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -31.00 |
| Energy contribution | -30.88 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11589953 120 - 20766785 GAACGUAAUUACACUCAGCCAGUUGUCCAAUCGAACGACGAAUAUCUGGGCGGGAUAACACAUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCCAAGCCAACCG ....((((((((((((((....(((((.........)))))....))))).(......)...)))))))))........((((..(((((((....)))))))..))))........... ( -31.50) >DroSec_CAF1 23518 113 - 1 GAACGUAAUUACACUCAGCCAGUUGUCCAAUCGAACGACGAAUAUCUGGGCGGGAUAACACAUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCUAAG------- ....((((((((((((((....(((((.........)))))....))))).(......)...)))))))))........((((..(((((((....)))))))..))))....------- ( -31.90) >DroSim_CAF1 26802 113 - 1 GAACGUAAUUACACUCAGCCAGUUGUCCAAUCGAACGACGAAUAUCUGGGCGGGAUAACACAUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCUAAG------- ....((((((((((((((....(((((.........)))))....))))).(......)...)))))))))........((((..(((((((....)))))))..))))....------- ( -31.90) >DroEre_CAF1 24191 115 - 1 GAAAGUAAUUACACUCGGCCAGUUGUCCAAUCGAACGACGAAUAUCUGGGCGGGAUAACACAUGUAAUUGCCUCAAAUUGUCGCAGUGUCCACUUCUGGACACGCCGACCAAGCC----- ....(((((((((((((.(((((((((.........)))))....)))).))))........)))))))))........((((..(((((((....)))))))..))))......----- ( -36.70) >DroYak_CAF1 24310 120 - 1 GAAAGUAAUUACACUCAGCCAGUUGUCCAAUCGAACGACGAAUAUCUGGGCGGGAUAACACAUGUAAUUGCCUCAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCAGCUAAGCCAGUCA ....((((((((((((((....(((((.........)))))....))))).(......)...)))))))))...........((.(((((((....)))))))))............... ( -28.40) >consensus GAACGUAAUUACACUCAGCCAGUUGUCCAAUCGAACGACGAAUAUCUGGGCGGGAUAACACAUGUAAUUGCCACAAAUUGUCGCAGUGUCCAUUUCUGGACACGCCGGCUAAGCC_____ ....((((((((((((((....(((((.........)))))....))))).(......)...)))))))))........((((..(((((((....)))))))..))))........... (-31.00 = -30.88 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:38 2006