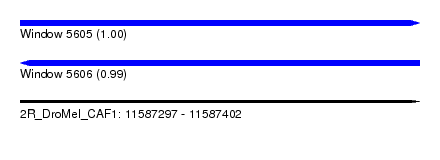
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,587,297 – 11,587,402 |
| Length | 105 |
| Max. P | 0.997738 |

| Location | 11,587,297 – 11,587,402 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -45.16 |
| Consensus MFE | -33.68 |
| Energy contribution | -33.56 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
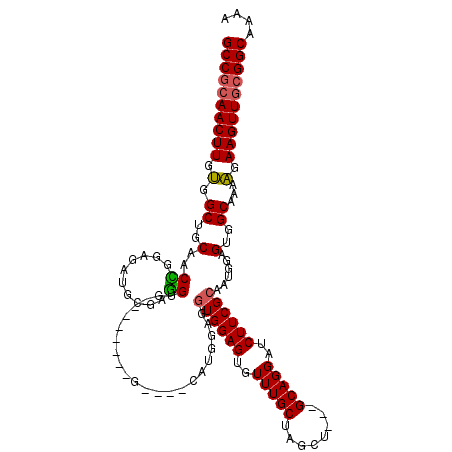
>2R_DroMel_CAF1 11587297 105 + 20766785 GCCGCAACUUGUGGCUGCAACCGGAGAUGCGUGGUG--------G----CAUGGCGAUGGAGUGUUUGCUAGCU---GCAGGAUCUUCGCAAUGGAGUGGCAAAAGAAGUUGCGGCAAAA ((((((((((((.(((.((..(((((((.(...(..--------(----(.((((((........)))))))).---.).).)))))))...)).))).))).....))))))))).... ( -48.30) >DroSec_CAF1 20860 108 + 1 GCCGCAACUUGUGGCUGCAACCGGAGAUGCGAGGUG--------G----CAUGGUGGUGGAGUGUUUGCUAGCUGCUGCAGGAUCUUCGCAAUGGAGUGGCAAAAGAAGUUACGGCAGAA ((((.(((((((.(((.((..(((((((.(...(..--------(----((.(.(((..((...))..))).))))..).).)))))))...)).))).))).....)))).)))).... ( -42.20) >DroSim_CAF1 24147 108 + 1 GCCGCAACUUGUGGCUGCAACCGGAGAUGCGUGGUG--------G----CAUGGUGGUGGAGUGUUUGCUAGCUGCUGCAGGAUCUUCGCAAUGGAGUGGCAAAAGAAGUUGCGGCAAAA ((((((((((((.(((.((..(((((((.((..(..--------(----(.(((..(........)..)))))..)..).).)))))))...)).))).))).....))))))))).... ( -49.60) >DroEre_CAF1 21712 102 + 1 GCCGCAACUUGCGGCUGCAACUGGAGAUGCGAGGCG---------------AGGAGGUGGAGUGUUUGCGGGCU---GCAGGAUCUUCGCAAUGGAGUGGCAAAGGAAGUUGCGGCAAAA ((((((((((.(.((..(..((.(.....).))(((---------------((((.(..(..((....))..).---.)....)))))))......)..))...).)))))))))).... ( -44.20) >DroYak_CAF1 21634 117 + 1 GCCACAACUUGUGGCUGCAACUGGAGAUGCGAAGCGUGCAGGUGGUUGAGGGGGAGGUGGAGUGUUUGCUAGCU---GCAGGAUCUUCGCAAUGGAGUGGCAAAAGAAGUUGCGGCAAAA (((.((((((.(.((..(..(......((((((((.(((((.((((.((.(...........).)).)))).))---))).)..)))))))..)..)..))...).)))))).))).... ( -41.50) >consensus GCCGCAACUUGUGGCUGCAACCGGAGAUGCGAGGUG________G____CAUGGAGGUGGAGUGUUUGCUAGCU___GCAGGAUCUUCGCAAUGGAGUGGCAAAAGAAGUUGCGGCAAAA ((((((((((.(.((..(..((..........))......................((((((..(((((........)))))..))))))......)..))...).)))))))))).... (-33.68 = -33.56 + -0.12)


| Location | 11,587,297 – 11,587,402 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
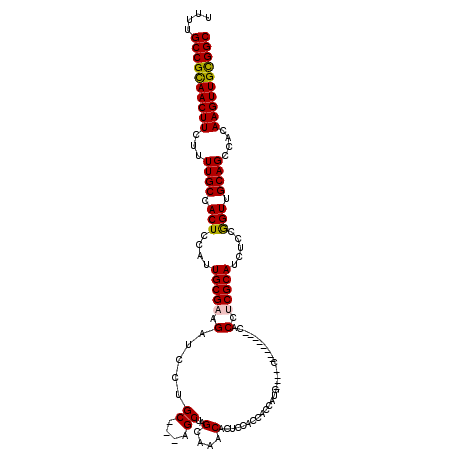
>2R_DroMel_CAF1 11587297 105 - 20766785 UUUUGCCGCAACUUCUUUUGCCACUCCAUUGCGAAGAUCCUGC---AGCUAGCAAACACUCCAUCGCCAUG----C--------CACCACGCAUCUCCGGUUGCAGCCACAAGUUGCGGC ....((((((((((.((((((.........))))))...((((---((((.((............)).(((----(--------......))))....))))))))....)))))))))) ( -31.10) >DroSec_CAF1 20860 108 - 1 UUCUGCCGUAACUUCUUUUGCCACUCCAUUGCGAAGAUCCUGCAGCAGCUAGCAAACACUCCACCACCAUG----C--------CACCUCGCAUCUCCGGUUGCAGCCACAAGUUGCGGC ....((((((((((.((((((.........))))))...(((((((.....)).........(((...(((----(--------......))))....))))))))....)))))))))) ( -28.00) >DroSim_CAF1 24147 108 - 1 UUUUGCCGCAACUUCUUUUGCCACUCCAUUGCGAAGAUCCUGCAGCAGCUAGCAAACACUCCACCACCAUG----C--------CACCACGCAUCUCCGGUUGCAGCCACAAGUUGCGGC ....((((((((((.((((((.........))))))...(((((((.....)).........(((...(((----(--------......))))....))))))))....)))))))))) ( -30.00) >DroEre_CAF1 21712 102 - 1 UUUUGCCGCAACUUCCUUUGCCACUCCAUUGCGAAGAUCCUGC---AGCCCGCAAACACUCCACCUCCU---------------CGCCUCGCAUCUCCAGUUGCAGCCGCAAGUUGCGGC ....((((((((((.((((((.........))))))...((((---(((..(....)............---------------.((...)).......)))))))....)))))))))) ( -28.00) >DroYak_CAF1 21634 117 - 1 UUUUGCCGCAACUUCUUUUGCCACUCCAUUGCGAAGAUCCUGC---AGCUAGCAAACACUCCACCUCCCCCUCAACCACCUGCACGCUUCGCAUCUCCAGUUGCAGCCACAAGUUGUGGC ....((((((((((...((((.(((....(((((((....(((---((...............................)))))..))))))).....))).))))....)))))))))) ( -33.78) >consensus UUUUGCCGCAACUUCUUUUGCCACUCCAUUGCGAAGAUCCUGC___AGCUAGCAAACACUCCACCACCAUG____C________CACCUCGCAUCUCCGGUUGCAGCCACAAGUUGCGGC ....((((((((((...((((.(((....(((((.(.....((....))..(....).............................).))))).....))).))))....)))))))))) (-23.74 = -23.58 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:26 2006