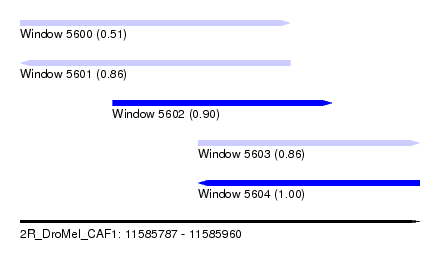
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,585,787 – 11,585,960 |
| Length | 173 |
| Max. P | 0.998513 |

| Location | 11,585,787 – 11,585,904 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.44 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11585787 117 + 20766785 GAUGGAAAAUCGAGGGCGGGCAGCACUUGACCUCCUGUGAGGCA---UAAAAAAAAUCGGAACCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACAC (((.....)))..(((..((((.(((..(....)..))).(((.---...........((......))(((((((.....))))))))))..)))).))).................... ( -32.70) >DroSec_CAF1 19345 115 + 1 GAUGGAAAAUCGAGGGCGGGCAGCACUUGACCUCCUGUGCGGCA---CAA--AAAAUCGGAACGAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACAC (((.....)))..(((..((((((((..(....)..))))(((.---...--......((......))(((((((.....))))))))))..)))).))).................... ( -36.80) >DroSim_CAF1 22157 115 + 1 GAUGGAAAAACGAGGGCGGGCAGCACUUGACCUCCUGUGCGGCA---CAA--AAAAUCGGAAUCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACAC .............(((..((((((((..(....)..))))(((.---...--......((......))(((((((.....))))))))))..)))).))).................... ( -35.80) >DroEre_CAF1 20229 114 + 1 GAUGGAAAAUCGAGGGCGGGCAGCACUUGACCUCCUGCGUGGCA---CAA--AAAAUCGGAGCCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACA- (((.....)))..(((.(((..(((.(((.......((.((((.---(..--......)..))))))((((((((.....)))))))).))))))..))).)))...............- ( -34.20) >DroYak_CAF1 20190 113 + 1 GAUGGAAAAUCGAGGGCGGGCAGCACUUGACCUCCUGUGAGGCACCACAA--AAAAUCGCAACCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACAC----- (((.....)))..(((.(((..(((..((.((((....))))))......--.............(.((((((((.....)))))))).)..)))..))).)))...........----- ( -32.10) >consensus GAUGGAAAAUCGAGGGCGGGCAGCACUUGACCUCCUGUGAGGCA___CAA__AAAAUCGGAACCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACAC (((.....)))..(((..((((((((..(....)..))))(((...............((......))(((((((.....))))))))))..)))).))).................... (-30.52 = -31.44 + 0.92)

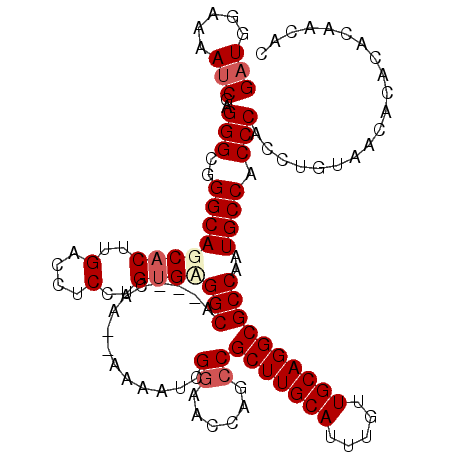
| Location | 11,585,787 – 11,585,904 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -36.33 |
| Energy contribution | -36.53 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11585787 117 - 20766785 GUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCAACAAAUGCAAGCGGCUGGUUCCGAUUUUUUUUA---UGCCUCACAGGAGGUCAAGUGCUGCCCGCCCUCGAUUUUCCAUC ....((((.....))))((((((..((((((.(((.((((.....)))).))).)..................---.(((((....)))))..)))))..)))))).............. ( -40.10) >DroSec_CAF1 19345 115 - 1 GUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCAACAAAUGCAAGCGGCUCGUUCCGAUUUU--UUG---UGCCGCACAGGAGGUCAAGUGCUGCCCGCCCUCGAUUUUCCAUC ....((((.....))))((((((..((((((.(((.((((.....)))).))).).......((((((--(((---((...))))))))))).)))))..)))))).............. ( -44.20) >DroSim_CAF1 22157 115 - 1 GUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCAACAAAUGCAAGCGGCUGAUUCCGAUUUU--UUG---UGCCGCACAGGAGGUCAAGUGCUGCCCGCCCUCGUUUUUCCAUC ....((((.....))))((((((..((((((.(((.((((.....)))).))).).......((((((--(((---((...))))))))))).)))))..)))))).............. ( -44.20) >DroEre_CAF1 20229 114 - 1 -UGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCAACAAAUGCAAGCGGCUGGCUCCGAUUUU--UUG---UGCCACGCAGGAGGUCAAGUGCUGCCCGCCCUCGAUUUUCCAUC -...((((.....))))((((((..(((((((.(((.((..(........)..)).))).))((((((--(((---((...))))))))))).)))))..)))))).............. ( -44.80) >DroYak_CAF1 20190 113 - 1 -----GUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCAACAAAUGCAAGCGGCUGGUUGCGAUUUU--UUGUGGUGCCUCACAGGAGGUCAAGUGCUGCCCGCCCUCGAUUUUCCAUC -----(((.....))).((((((..((((((.(((.((((.....)))).))).).......((((((--((((((....)))))))))))).)))))..)))))).............. ( -45.20) >consensus GUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCAACAAAUGCAAGCGGCUGGUUCCGAUUUU__UUG___UGCCGCACAGGAGGUCAAGUGCUGCCCGCCCUCGAUUUUCCAUC ....((((.....))))((((((..((((((((((.((((.....)))).))(((......................))).........))).)))))..)))))).............. (-36.33 = -36.53 + 0.20)

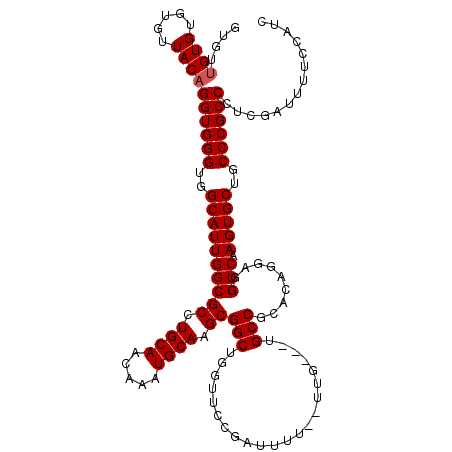
| Location | 11,585,827 – 11,585,922 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11585827 95 + 20766785 GGCA---UAAAAAAAAUCGGAACCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACU----------------------GCGUGUG ((((---(..........(....).(.((((((((.....)))))))).).)))))............(((((.((..............)----------------------).))))) ( -25.14) >DroSec_CAF1 19385 93 + 1 GGCA---CAA--AAAAUCGGAACGAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACU----------------------GCGUGUG ((((---...--....(((...)))(.((((((((.....)))))))).)..))))............(((((.((..............)----------------------).))))) ( -24.24) >DroSim_CAF1 22197 93 + 1 GGCA---CAA--AAAAUCGGAAUCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACU----------------------GCGUGUG ((((---...--......((......))(((((((.....))))))).....))))............(((((.((..............)----------------------).))))) ( -23.84) >DroEre_CAF1 20269 113 + 1 GGCA---CAA--AAAAUCGGAGCCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACA--CAACCACACUCUGCACGGCUGUAUCUAAGCGUGUGUGUG ((((---...--......(....).(.((((((((.....)))))))).)..))))...........((((((((....--....((.....))....(((.......))))))))))). ( -31.70) >DroYak_CAF1 20230 80 + 1 GGCACCACAA--AAAAUCGCAACCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACAC--------------------------------------GGGUGUG ((((......--......((.....))((((((((.....))))))))....))))...(((((((.......))--------------------------------------))))).. ( -27.50) >consensus GGCA___CAA__AAAAUCGGAACCAGCCGCUUGCAUUUGUUGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACU______________________GCGUGUG (((...............((......))(((((((.....))))))))))..................(((((..........................................))))) (-19.67 = -19.87 + 0.20)

| Location | 11,585,864 – 11,585,960 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -23.59 |
| Energy contribution | -26.15 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11585864 96 + 20766785 UGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACUGCGUGUGUAUGU--GUGUAUGAAUUGGAUAUGCGGCGGGGGUGGGUG ....((((....))))((((((((.(..((((((((((((((...........)))))).)))--)))).......(......))..).)))))))). ( -35.10) >DroSec_CAF1 19420 91 + 1 UGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACUGCGUGUGUAUGUGUGUGUAUGGAUUGGAUAUGCGGCGGGG------- ....((((....)))).(((..((((((((((((((((((((...........)))))).)))))))))............)))))..)))------- ( -33.10) >DroSim_CAF1 22232 91 + 1 UGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACUGCGUGUGUAUGUGUGUGUAUGGACUGGAUAUGCGGCGGGG------- ....((((....)))).(((..((((((((((((((((((((...........)))))).)))))))))............)))))..)))------- ( -33.10) >DroYak_CAF1 20268 81 + 1 UGCAGGCGCCAAUGCCACCCACCUGUAACACACAC----------------GGGUGUGUGUGUGUGUGUAUGGAUUGGAUACACGUGGGGG-UGGUGG ......((((...(((.(((((.....((((((((----------------(....)))))))))((((((.......)))))))))))))-))))). ( -34.80) >consensus UGCAGGCGCCAAUGCCACCCACCUGUAACACACACAACACACAACCGCACUGCGUGUGUAUGUGUGUGUAUGGAUUGGAUAUGCGGCGGGG_______ ....((((....)))).(((..((((((((((((((((((((...........)))))).)))))))))............)))))..)))....... (-23.59 = -26.15 + 2.56)

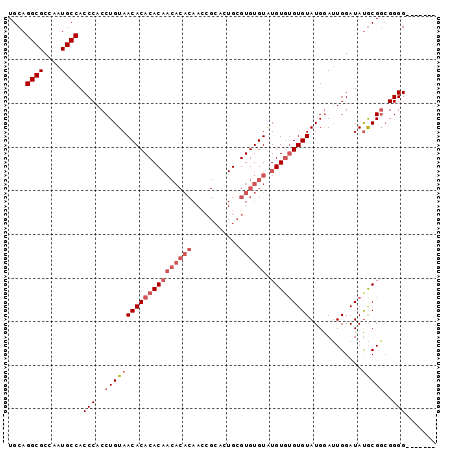
| Location | 11,585,864 – 11,585,960 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -29.75 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11585864 96 - 20766785 CACCCACCCCCGCCGCAUAUCCAAUUCAUACAC--ACAUACACACGCAGUGCGGUUGUGUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCA ((((((((......(......)....(((((((--(((((((..(((...)))..)))))).)))))))).....))))))))(((........))). ( -35.70) >DroSec_CAF1 19420 91 - 1 -------CCCCGCCGCAUAUCCAAUCCAUACACACACAUACACACGCAGUGCGGUUGUGUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCA -------...((((((.((((((......((((((((((((((((((...)))..)))))).)))))))))......))))))))...))))...... ( -36.50) >DroSim_CAF1 22232 91 - 1 -------CCCCGCCGCAUAUCCAGUCCAUACACACACAUACACACGCAGUGCGGUUGUGUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCA -------...((((((.((((((.(....((((((((((((((((((...)))..)))))).)))))))))....).))))))))...))))...... ( -36.20) >DroYak_CAF1 20268 81 - 1 CCACCA-CCCCCACGUGUAUCCAAUCCAUACACACACACACACACCC----------------GUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCA ((((((-(((((..((((((.......))))))((((((((......----------------))))))))....)).))))))...)))........ ( -32.60) >consensus _______CCCCGCCGCAUAUCCAAUCCAUACACACACAUACACACGCAGUGCGGUUGUGUGUUGUGUGUGUUACAGGUGGGUGGCAUUGGCGCCUGCA ..............(((...((((((((..(((......((((((((((..((....))..)))))))))).....)))..))).)))))....))). (-29.75 = -29.50 + -0.25)


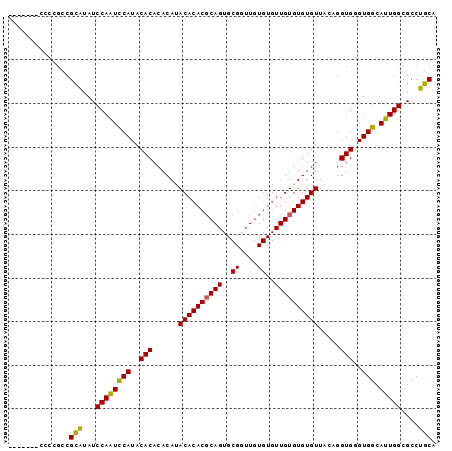
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:24 2006