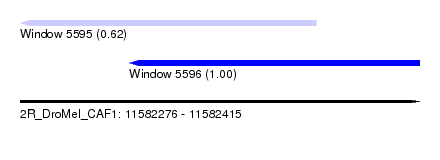
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,582,276 – 11,582,415 |
| Length | 139 |
| Max. P | 0.996605 |

| Location | 11,582,276 – 11,582,379 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -11.05 |
| Energy contribution | -12.77 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11582276 103 - 20766785 CU---------UCUGUUAGCCAACUUGAGUGGAAUUCCCAA-UG-GAAUUCUAUGGGGCAAUCAAAGUGGGCAGCU------AACCCAUCGCAUCGACAGGGGUAAAUAUUUUAGGAAAU ((---------((((((.(((.......((((((((((...-.)-)))))))))..))).......(((((.....------..)))))......))))))))................. ( -29.40) >DroSec_CAF1 16012 112 - 1 CUCC-------UCUGUUGGCCAACUUGAGUGGAAUUCCCAUGUG-GAAUUCUAUGGGGCAAUCAAAGUGGGCAACUGCUACUAACCCAACGCAUCGACAGGGGUAAAUAUUUUAGGAAAA ..((-------((((((((((.......((((((((((.....)-)))))))))..)))......((..(....)..))...............)))))))))................. ( -35.50) >DroSim_CAF1 16815 113 - 1 CUCC-------UCUGUUGGCCAACUUGAGUGGAAUUCCUAUGUGGGAAUUCUAUGGGGCAAUCAAAGUGGGCAACUACUACCAACCCAACGCAUCGACAGGGGUAAAUAUUUUAGGAAAU ..((-------((((((((((.......(((((((((((....)))))))))))..)))......(((((....)))))...............)))))))))................. ( -38.20) >DroEre_CAF1 16245 91 - 1 C---------UCCUAUCAGCCAACUUGAGUGGAAUUCCUU--------CUCUAUGG-GCAAUCAAAGUGGGCAUCU-----------AUCCCAUCGACGCGGGUAAAUAUUUUAGGAUAU .---------(((((...(((.(((((((.((....))..--------)))..(((-....))))))).)))...(-----------(((((......).))))).......)))))... ( -20.90) >DroYak_CAF1 16598 101 - 1 CUCCAAUCUAUCCCAUCAGCCAACUUGAGUGGCAUUCCUU--------CUCUAUGGGGCAAUCAAAGUGGGCACCU-----------AUCCCAUGGACAUGGGUAAAUAUUUUCGGAAAU .(((((..((((((((..((((.(....))))).......--------.((((((((.........((((....))-----------)))))))))).)))))...)))..)).)))... ( -24.80) >consensus CUCC_______UCUGUUAGCCAACUUGAGUGGAAUUCCUA__UG_GAAUUCUAUGGGGCAAUCAAAGUGGGCAACU______AACCCAUCGCAUCGACAGGGGUAAAUAUUUUAGGAAAU ...........((((...(((.(((((((((((((((........))))))))).......)).)))).)))....................(((......)))........)))).... (-11.05 = -12.77 + 1.72)


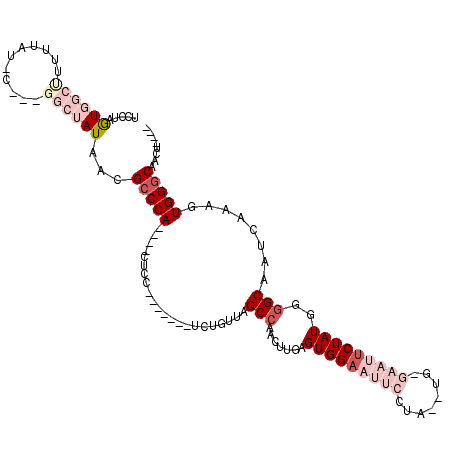
| Location | 11,582,314 – 11,582,415 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -11.61 |
| Energy contribution | -15.17 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11582314 101 - 20766785 UCCUAGUGGCUUUUUAUACGCUGGCUAUAACGCCCA----CU---------UCUGUUAGCCAACUUGAGUGGAAUUCCCAA-UG-GAAUUCUAUGGGGCAAUCAAAGUGGGCAGCU---- ..((((((..........)))))).......(((((----((---------(..(((.(((.......((((((((((...-.)-)))))))))..))))))..))))))))....---- ( -37.60) >DroSec_CAF1 16052 96 - 1 UCCUAGUGGCUU-UU-----------AUAACGCCCA----CUCC-------UCUGUUGGCCAACUUGAGUGGAAUUCCCAUGUG-GAAUUCUAUGGGGCAAUCAAAGUGGGCAACUGCUA .((....))...-..-----------.....(((((----((..-------...((((.((.......((((((((((.....)-))))))))).)).))))...)))))))........ ( -30.60) >DroSim_CAF1 16855 98 - 1 UCCUAGUGGCUUUUU-----------AUAACGCCCA----CUCC-------UCUGUUGGCCAACUUGAGUGGAAUUCCUAUGUGGGAAUUCUAUGGGGCAAUCAAAGUGGGCAACUACUA ...((((((..((..-----------..)).(((((----((..-------...((((.((.......(((((((((((....))))))))))).)).))))...)))))))..)))))) ( -33.10) >DroEre_CAF1 16278 91 - 1 UCCCCAUGGCCUUUUAUAC---GGCUAUAACGGCCA----C---------UCCUAUCAGCCAACUUGAGUGGAAUUCCUU--------CUCUAUGG-GCAAUCAAAGUGGGCAUCU---- .....((((((........---))))))...(.(((----(---------(.......(((.....(((.((....))..--------)))....)-))......))))).)....---- ( -25.72) >DroYak_CAF1 16631 105 - 1 UCCUCGUGGCCUUUUAUGC---GGCUAUAACGCCCAACCACUCCAAUCUAUCCCAUCAGCCAACUUGAGUGGCAUUCCUU--------CUCUAUGGGGCAAUCAAAGUGGGCACCU---- .....((((((........---))))))...(((((.((((((.......................))))))...((((.--------......)))).........)))))....---- ( -25.90) >consensus UCCUAGUGGCUUUUUAU_C___GGCUAUAACGCCCA____CUCC_______UCUGUUAGCCAACUUGAGUGGAAUUCCUA__UG_GAAUUCUAUGGGGCAAUCAAAGUGGGCAACU____ .....((((((...........))))))...(((((......................(((.......(((((((((........)))))))))..)))........)))))........ (-11.61 = -15.17 + 3.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:17 2006