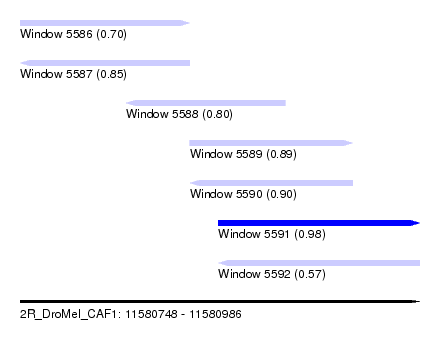
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,580,748 – 11,580,986 |
| Length | 238 |
| Max. P | 0.980008 |

| Location | 11,580,748 – 11,580,849 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -23.18 |
| Energy contribution | -25.12 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580748 101 + 20766785 ---U---G-GGUGGUGUUUGUAAUGGACCACCACCCAUGU---GGCAGGCCACAAACGAGCAUCACUUUCAAUGAAAUUAGCUUGUUUCGCAAUCGUUCUUGCCACCUAGC-- ---(---(-((((((((((.....))).))))))))).((---((((((..((((((((((.....((((...))))...)))))))).......)).)))))))).....-- ( -33.41) >DroSec_CAF1 14515 109 + 1 UGUU---G-GGUGGUGUUUGUAAUGGACCACCACCCAUGUGGUGGCAGGCCACAAACGAGCAUCACUUUCAAUGAAAUUAGCUUGUUUCGCAAUCGUUCUUGCCACCUAGCGC .(((---(-((((((((((((...((.(((((((....)))))))....))))))))((((.(((.......))).....((.......))....))))..)))))))))).. ( -39.40) >DroSim_CAF1 15313 102 + 1 -----------UGGUGUUUGUAAUGGACCACCACCCAUGUGGUGGCAGGCCACAAACGAGCAUCACUUUCAAUGAAAUUAGCUUGUUUCGCAAUCGUUCUUGCCACCUAGCGC -----------((((((((((..(((.(((((((....)))))))....)))...))))))))))...............(((......((((......)))).....))).. ( -30.60) >DroEre_CAF1 14742 97 + 1 -------GGGGAGGUGCCUGUAAUGGACC---ACCCAUGU---GGCAGGCCACAAACGAGCAUCACUUUCAAUGAAAUUAGCUUGUUUCGCAAUCGUUCUUGCCACCUCG--- -------((((.(((((((((.((((...---..))))..---.))))))...((((((((.....((((...))))...)))))))).............))).)))).--- ( -31.10) >DroYak_CAF1 15025 106 + 1 UAUUGAGGGGUGGGUGUUUGUAAUGGACC---ACCCAUGU---GGCAGGCCACAAACGAGCAUCACUUUCAAUGAAAUUAGCUUGUUUCGCAAUCGUUCUUGCCACCUCC-GC ....(.(((((((((((((((.((((...---..))))((---((....))))..))))))))).........(((((......)))))((((......)))))))))))-.. ( -33.70) >consensus ___U___G_GGUGGUGUUUGUAAUGGACCACCACCCAUGU___GGCAGGCCACAAACGAGCAUCACUUUCAAUGAAAUUAGCUUGUUUCGCAAUCGUUCUUGCCACCUAGCGC .........((((((((((((..(((.(((((((....)))))))....)))...))))))))))))......(((((......)))))((((......)))).......... (-23.18 = -25.12 + 1.94)


| Location | 11,580,748 – 11,580,849 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580748 101 - 20766785 --GCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUCAUUGAAAGUGAUGCUCGUUUGUGGCCUGCC---ACAUGGGUGGUGGUCCAUUACAAACACCACC-C---A--- --....((((((..((((((..((.......)).....((((.....))))...))))))...))).)))---...(((((((((............)))))))-)---)--- ( -34.20) >DroSec_CAF1 14515 109 - 1 GCGCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUCAUUGAAAGUGAUGCUCGUUUGUGGCCUGCCACCACAUGGGUGGUGGUCCAUUACAAACACCACC-C---AACA ......(((((((.(..((...((((.....((.....((((.....)))).))))))...))..))))))))...(((((((((............)))))))-)---)... ( -37.90) >DroSim_CAF1 15313 102 - 1 GCGCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUCAUUGAAAGUGAUGCUCGUUUGUGGCCUGCCACCACAUGGGUGGUGGUCCAUUACAAACACCA----------- ......((((((((......)))).((((.(((.....((((.....)))).))).))))((((.(..(((((.....)))))..).))))......)))).----------- ( -34.00) >DroEre_CAF1 14742 97 - 1 ---CGAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUCAUUGAAAGUGAUGCUCGUUUGUGGCCUGCC---ACAUGGGU---GGUCCAUUACAGGCACCUCCCC------- ---.((((((((((......)))).((((.(((.....((((.....)))).))).))))((((.(..((---.....)).---.).))))......))))))...------- ( -29.80) >DroYak_CAF1 15025 106 - 1 GC-GGAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUCAUUGAAAGUGAUGCUCGUUUGUGGCCUGCC---ACAUGGGU---GGUCCAUUACAAACACCCACCCCUCAAUA ..-.((((.(((((......)))).((((.(((.....((((.....)))).))).))))((((....))---)).(((((---(............))))))).)))).... ( -30.90) >consensus GCGCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUCAUUGAAAGUGAUGCUCGUUUGUGGCCUGCC___ACAUGGGUGGUGGUCCAUUACAAACACCACC_C___A___ ......((((((((......)))).((((.(((.....((((.....)))).))).))))((((...((((((((....))))))))))))......))))............ (-26.04 = -26.94 + 0.90)

| Location | 11,580,811 – 11,580,906 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.54 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580811 95 - 20766785 GCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA--------------------GAGGCAACCGAGGUG-----GCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUC ((((.((((((((((...((((....(..((((....)))).--------------------.).))))....))))-----)).)))).((((......)))).....))))....... ( -30.80) >DroSec_CAF1 14584 100 - 1 GCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA--------------------GAGGCAGACGAGGCAGUGGCGCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUC ((((((.(((((.....(((....)))..((((....)))).--------------------..))))).))))))..((((......(.((((......)))).).....))))..... ( -29.40) >DroSim_CAF1 15375 100 - 1 GCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA--------------------GAGGCAGACGAGGCAGUGGCGCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUC ((((((.(((((.....(((....)))..((((....)))).--------------------..))))).))))))..((((......(.((((......)))).).....))))..... ( -29.40) >DroEre_CAF1 14802 94 - 1 GCUUUGCCUGCCACCAAAUUGCUAAAUAAUUGCCACGGCAA---------------------GAGGCAGCCGAGGUGG-----CGAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUC ((((.((((((((((...(((((......((((....))))---------------------..)))))....)))))-----).)))).((((......)))).....))))....... ( -31.50) >DroYak_CAF1 15092 119 - 1 GCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAAGAGGCAAGCUGAGGCAAGCGGAGGCAGCUGAGGCACCAGC-GGAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUC (((((..(((((.((...((((.......(((((.(......).)))))......))))..)).)))))..)))))...(((-.....(.((((......)))).).....)))...... ( -38.52) >consensus GCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA____________________GAGGCAGCCGAGGCAG__GCGCUAGGUGGCAAGAACGAUUGCGAAACAAGCUAAUUUC ((((((.(((((.....(((....)))..((((....)))).......................))))).))))))..............((((......))))................ (-22.74 = -22.54 + -0.20)

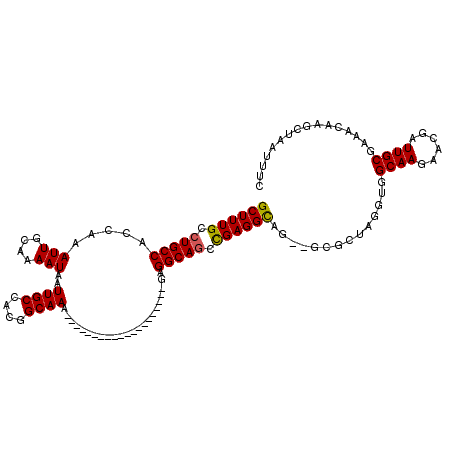
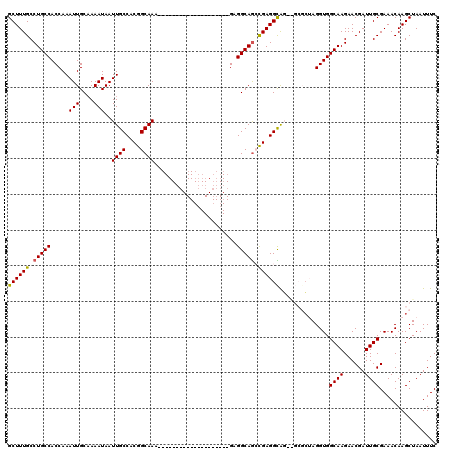
| Location | 11,580,849 – 11,580,946 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -23.37 |
| Energy contribution | -23.81 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580849 97 + 20766785 ---CACCUCGGUUGCCUC--------------------UUUGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCAA ---...(((..((((((.--------------------.(..(((..((((.....))))....)))..).)))))).(((((.........)))))........))).((.....)).. ( -27.60) >DroSec_CAF1 14624 100 + 1 CACUGCCUCGUCUGCCUC--------------------UUUGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGA ...(((((((((((((..--------------------...((((..((((.....))))....)))))))))))...(((((.........)))))........))).)))........ ( -33.10) >DroSim_CAF1 15415 100 + 1 CACUGCCUCGUCUGCCUC--------------------UUUGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGA ...(((((((((((((..--------------------...((((..((((.....))))....)))))))))))...(((((.........)))))........))).)))........ ( -33.10) >DroEre_CAF1 14839 97 + 1 --CCACCUCGGCUGCCUC---------------------UUGCCGUGGCAAUUAUUUAGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGA --....(((...(((((.---------------------(..(((..((.........))....)))..).)))))..(((((.........)))))........)))(((.....))). ( -26.90) >DroYak_CAF1 15131 120 + 1 UGGUGCCUCAGCUGCCUCCGCUUGCCUCAGCUUGCCUCUUUGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGA .(((((....)).)))..((((((((((.(((((((.....((((..((((.....))))....)))))))))))...(((((.........)))))........))).))))...))). ( -40.40) >consensus __CUGCCUCGGCUGCCUC____________________UUUGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGA ......((((.(((((.........................((((..((((.....))))....))))))))).)...(((((.........)))))........)))(((.....))). (-23.37 = -23.81 + 0.44)

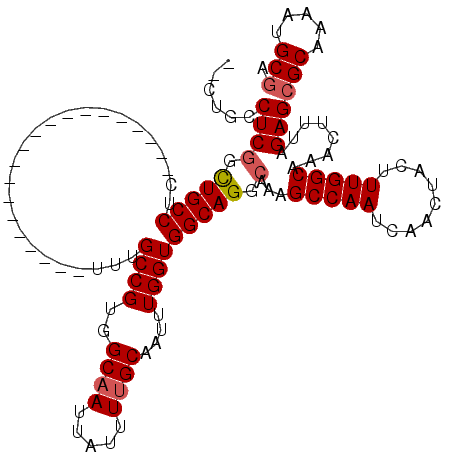
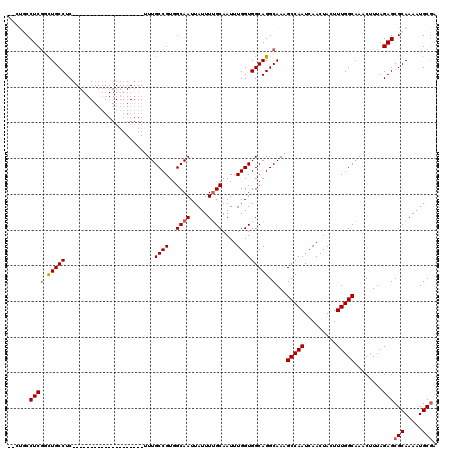
| Location | 11,580,849 – 11,580,946 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -27.28 |
| Energy contribution | -26.72 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580849 97 - 20766785 UUGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA--------------------GAGGCAACCGAGGUG--- ..((.....))((.((...(.(((((...((((((...((((.......))))...)))))).......((((....)))).--------------------..))))).).)))).--- ( -25.80) >DroSec_CAF1 14624 100 - 1 UCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA--------------------GAGGCAGACGAGGCAGUG ..((.....))((.((...(((((((...((((((...((((.......))))...)))))).......((((....)))).--------------------..))))))).)))).... ( -32.80) >DroSim_CAF1 15415 100 - 1 UCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA--------------------GAGGCAGACGAGGCAGUG ..((.....))((.((...(((((((...((((((...((((.......))))...)))))).......((((....)))).--------------------..))))))).)))).... ( -32.80) >DroEre_CAF1 14839 97 - 1 UCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCUAAAUAAUUGCCACGGCAA---------------------GAGGCAGCCGAGGUGG-- ...(((((((.(((.........(((..(((((((...((((.......))))...)))))))......((((....))))---------------------..))))))))))))).-- ( -28.40) >DroYak_CAF1 15131 120 - 1 UCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAAGAGGCAAGCUGAGGCAAGCGGAGGCAGCUGAGGCACCA ((((...((((.(((...((((((((...((((((...((((.......))))...)))))).......((((....))))...))))))))))))))))))).((..((....)).)). ( -40.70) >consensus UCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCAAA____________________GAGGCAGCCGAGGCAG__ ..((.....))((.((...(.(((((...((((((...((((.......))))...)))))).......((((....)))).......................))))).).)))).... (-27.28 = -26.72 + -0.56)

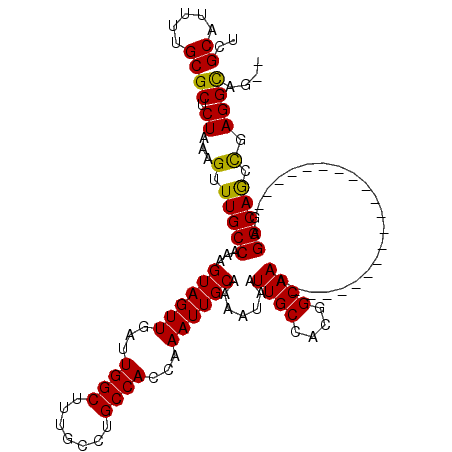
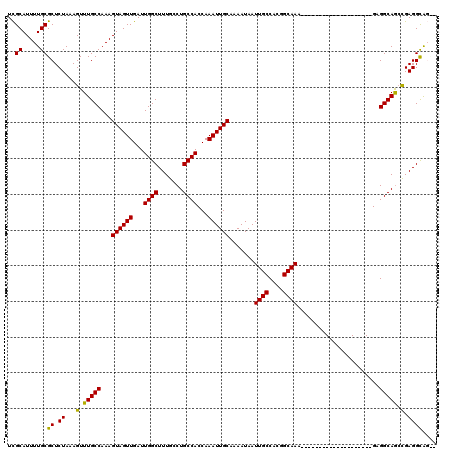
| Location | 11,580,866 – 11,580,986 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580866 120 + 20766785 UGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCAAAAUGUAAAUAAAAUUAAGUCCCCUGCCAUGGGGGGGUAGC ((((...((((.....))))..(((.((((((((....(((((.........)))))............((.....)).......................)))))))).))).)))).. ( -31.50) >DroSec_CAF1 14644 116 + 1 UGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGAAAUGUAAAUAAAAUGAAGUCCCCUGCCAU----GGGUAGC ((((...((((.....))))......((((((((....(((((.........)))))..((((((...(((.....)))..............))))))..))))))))----.)))).. ( -33.33) >DroSim_CAF1 15435 116 + 1 UGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGAAAUGUAAAUAAAAUUAAGUCCCCUGCCAG----GGGUAGC ((((...((((.....)))).......(((((((....(((((.........)))))....((((.(.(((.....)))...).)))).............))))))).----.)))).. ( -31.50) >DroEre_CAF1 14856 116 + 1 UGCCGUGGCAAUUAUUUAGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGAAAUGUAAAUAAAAUUAAGUCCCCUGCCAU----GGCAGGC ((((((((((........((......))((..(((...(((((.........)))))....((((.(.(((.....)))...).)))).........))).))))))))----))))... ( -32.00) >DroYak_CAF1 15171 116 + 1 UGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGAAAUGUAAAUAAAAUUAAGUCCCCUGCCAU----GGCAGGC ((((...((((.....))))......((((((((....(((((.........)))))....((((.(.(((.....)))...).)))).............))))))))----))))... ( -35.00) >consensus UGCCGUGGCAAUUAUUUUGCAAUUUGGUGGCAGGCAAAGCCAAUCAACUACUUUGGCAAACUUUAGAGCGCAAAAUGCGAAAUGUAAAUAAAAUUAAGUCCCCUGCCAU____GGGUAGC .((((..((((.....))))....))))((((((....(((((.........)))))....((((.(.(((.....)))...).)))).............))))))............. (-28.52 = -28.92 + 0.40)

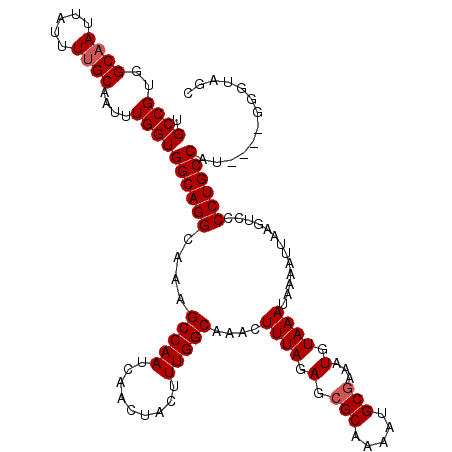

| Location | 11,580,866 – 11,580,986 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -26.98 |
| Energy contribution | -26.78 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11580866 120 - 20766785 GCUACCCCCCCAUGGCAGGGGACUUAAUUUUAUUUACAUUUUGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCA .........((.((((((....)..............(((((((((((((.((..........))))))))((((...((((.......))))...)))))))))))...))))).)).. ( -28.50) >DroSec_CAF1 14644 116 - 1 GCUACCC----AUGGCAGGGGACUUCAUUUUAUUUACAUUUCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCA (((....----.(((((((.((..(((..(((.((((.(((.(((....((........)).))).))))))).))).)))..)).))))))).......((((.....))))...))). ( -27.60) >DroSim_CAF1 15435 116 - 1 GCUACCC----CUGGCAGGGGACUUAAUUUUAUUUACAUUUCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCA (((.(((----(.....)))).....................(((.((((.((..........))))))((((((...((((.......))))...))))))........)))...))). ( -26.70) >DroEre_CAF1 14856 116 - 1 GCCUGCC----AUGGCAGGGGACUUAAUUUUAUUUACAUUUCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCUAAAUAAUUGCCACGGCA ....(((----.((((((.((....(((((((.........(((.....)))...)))))))..))..(((((((...((((.......))))...)))))))......)))))).))). ( -31.10) >DroYak_CAF1 15171 116 - 1 GCCUGCC----AUGGCAGGGGACUUAAUUUUAUUUACAUUUCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCA ....(((----.((((((.((....(((((((.........(((.....)))...)))))))..))...((((((...((((.......))))...)))))).......)))))).))). ( -30.40) >consensus GCUACCC____AUGGCAGGGGACUUAAUUUUAUUUACAUUUCGCAUUUUGCGCUCUAAAGUUUGCCAAAGUAGUUGAUUGGCUUUGCCUGCCACCAAAUUGCAAAAUAAUUGCCACGGCA (((.........(((((((.((...................(((.....)))...........(((((.........))))).)).))))))).......((((.....))))...))). (-26.98 = -26.78 + -0.20)

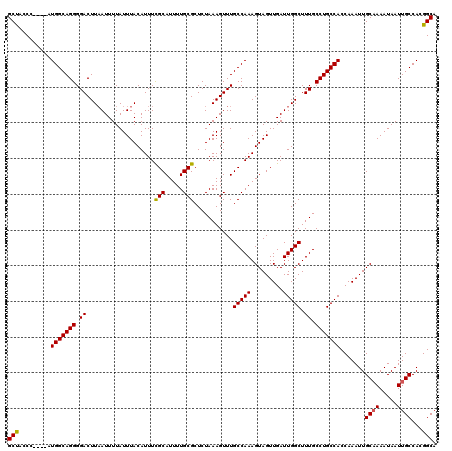
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:13 2006