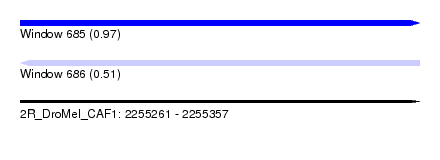
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,255,261 – 2,255,357 |
| Length | 96 |
| Max. P | 0.967198 |

| Location | 2,255,261 – 2,255,357 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -15.83 |
| Energy contribution | -17.03 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
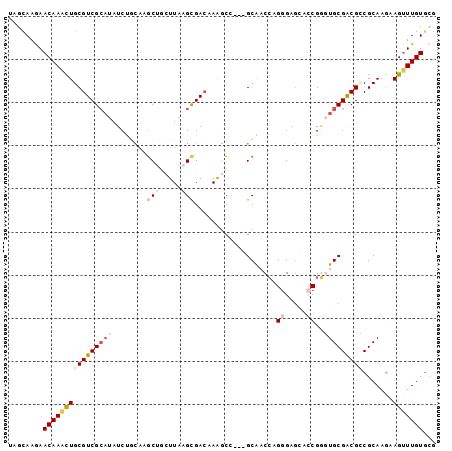
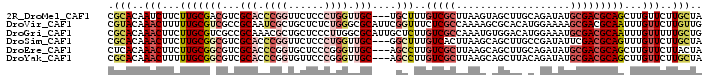
>2R_DroMel_CAF1 2255261 96 + 20766785 UAGCAAGAACAAGCUGCGUCGCAUAUCUGCAAGCUACUUAAGCGACAAAGCA---GCAACCAGGGAGAACCGGGUGCGACGUCGCAAGAAGAUUGUGCG ..(((.......((.(((((((((..((((..(((.....)))......)))---)...((.((.....))))))))))))).))..........))). ( -29.43) >DroVir_CAF1 39956 99 + 1 CAACAAGAACAAAUUGCGUCGCUUUUCCAUGUGCGCUUUUGGCGAGAAACCGAAUGCGCCCAGAGAGCAGCGAUUGCGGCGACGCAAAAAGUUUGUACG ......(.(((((((...(((((.(((..((.((((.(((((.......))))).)))).))..))).)))))(((((....)))))..))))))).). ( -35.50) >DroGri_CAF1 26996 99 + 1 CAGCAAAAACAAAUUGCGUCGCAUUUCCAUGUCCACAUUUGGCGACAAGAGCAAUGCGCCAAGGGAGCAGCGUUUGCGGCGACGCAAGAAGUUUGUGCG ..(((.((((...(((((((((..(((((((....)))((((((.((.......))))))))))))((((...)))).)))))))))...)))).))). ( -38.20) >DroSim_CAF1 22248 96 + 1 UAGCAAGAACAAACUGCGUCGAAUAUCGGCAAGCUGCUUAAGUGACAAAGCC---GCAACCAGGGAGAACCGGGUGCGACGCCGCAAGAAGUUUGUGCG ..((....((((((((((((.......(((..(((.....)))......)))---(((.((.((.....)).))))))))))(....).))))))))). ( -31.20) >DroEre_CAF1 22143 96 + 1 UAGUAAGAACAAGCUGCGUCGCAUAUCUGCAAGCUGCUUAAGCGACAAGGCU---GCAACCCGGGAGCACCGGGUGCGACGCCGCAAGAAGUUUGUGAG ........((((((((((((((.....((((.(((((....)).....))))---)))((((((.....)))))))))))))(....).)))))))... ( -40.50) >DroYak_CAF1 18294 96 + 1 UAGCAAGAACAAGCUGCGUCGCAUAUCUGUAAGCUGCUUAAGCGACAAGGCU---GCAACCCGGGAACACCGGGUGCGACGCCGCAAAAAGUUUGUGCG ..(((.((((..((.(((((((..........((.((((........)))).---)).((((((.....))))))))))))).)).....)))).))). ( -40.40) >consensus UAGCAAGAACAAACUGCGUCGCAUAUCUGCAAGCUGCUUAAGCGACAAAGCC___GCAACCAGGGAGCACCGGGUGCGACGCCGCAAGAAGUUUGUGCG ........((((((((((((((((........(((.....)))...................((.....))..))))))))).......)))))))... (-15.83 = -17.03 + 1.20)

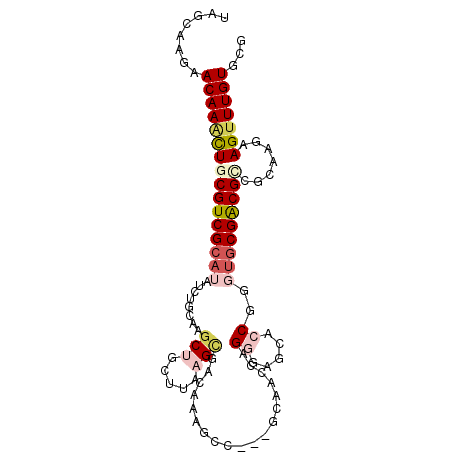
| Location | 2,255,261 – 2,255,357 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -13.48 |
| Energy contribution | -15.34 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.41 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2255261 96 - 20766785 CGCACAAUCUUCUUGCGACGUCGCACCCGGUUCUCCCUGGUUGC---UGCUUUGUCGCUUAAGUAGCUUGCAGAUAUGCGACGCAGCUUGUUCUUGCUA .(((..........(((((((.(((.((((......)))).)))---.))...))))).(((((.(((((((....))))).)).)))))....))).. ( -30.30) >DroVir_CAF1 39956 99 - 1 CGUACAAACUUUUUGCGUCGCCGCAAUCGCUGCUCUCUGGGCGCAUUCGGUUUCUCGCCAAAAGCGCACAUGGAAAAGCGACGCAAUUUGUUCUUGUUG ...((((((...(((((((((.(((.....)))(((.((.((((.((.(((.....))).)).)))).)).)))...)))))))))...))..)))).. ( -32.70) >DroGri_CAF1 26996 99 - 1 CGCACAAACUUCUUGCGUCGCCGCAAACGCUGCUCCCUUGGCGCAUUGCUCUUGUCGCCAAAUGUGGACAUGGAAAUGCGACGCAAUUUGUUUUUGCUG .(((.((((...(((((((((.(((.....)))(((.((((((((.......)).))))))(((....))))))...)))))))))...)))).))).. ( -36.30) >DroSim_CAF1 22248 96 - 1 CGCACAAACUUCUUGCGGCGUCGCACCCGGUUCUCCCUGGUUGC---GGCUUUGUCACUUAAGCAGCUUGCCGAUAUUCGACGCAGUUUGUUCUUGCUA .(((..(((.....((.((((((((.((((......)))).)))---(((.((((.......))))...))).......))))).))..)))..))).. ( -29.30) >DroEre_CAF1 22143 96 - 1 CUCACAAACUUCUUGCGGCGUCGCACCCGGUGCUCCCGGGUUGC---AGCCUUGUCGCUUAAGCAGCUUGCAGAUAUGCGACGCAGCUUGUUCUUACUA ..............((.(((((((((((((.....)))))((((---(((((((.....))))..)).)))))...)))))))).))............ ( -34.10) >DroYak_CAF1 18294 96 - 1 CGCACAAACUUUUUGCGGCGUCGCACCCGGUGUUCCCGGGUUGC---AGCCUUGUCGCUUAAGCAGCUUACAGAUAUGCGACGCAGCUUGUUCUUGCUA .(((..(((.....((.((((((((.((((.....))))(((((---(((......)))...))))).........)))))))).))..)))..))).. ( -35.90) >consensus CGCACAAACUUCUUGCGGCGUCGCACCCGGUGCUCCCUGGUUGC___AGCCUUGUCGCUUAAGCAGCUUACAGAUAUGCGACGCAGCUUGUUCUUGCUA .(((..(((...(((((((...(((.((((......)))).)))....)))..(((((...................)))))))))...)))..))).. (-13.48 = -15.34 + 1.86)
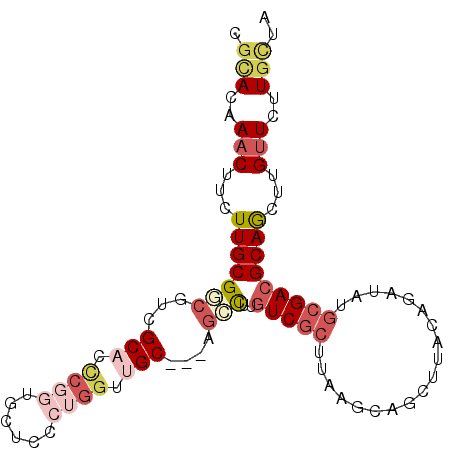
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:59 2006