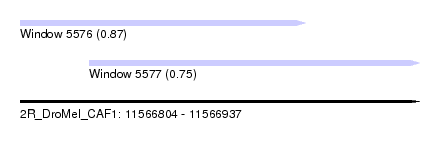
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,566,804 – 11,566,937 |
| Length | 133 |
| Max. P | 0.868935 |

| Location | 11,566,804 – 11,566,899 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.34 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11566804 95 + 20766785 GCUCCUCCUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGGAGCAAGCCGGC----UGG (((.(((((....(((.(((((((.((((((((((((((..........)))))))).)))----))))).))))))))...)))))..)))....----... ( -36.80) >DroSec_CAF1 3477 95 + 1 GCUCCUCUUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGAAGCAAGCCGGC----UGG (((...((((((((((.(((((((.((((((((((((((..........)))))))).)))----))))).)))))))).....)))))))..)))----... ( -36.60) >DroSim_CAF1 3619 95 + 1 GCUCCUCCUGCUUCCGUAUUUGCUCGCCUGUUUUUGUGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGGAGCAAGCCGGC----UGG (((.(((((....(((.(((((((.(((((((((((..(..........)..))))).)))----))))).))))))))...)))))..)))....----... ( -34.30) >DroEre_CAF1 3605 93 + 1 GCUCC--CUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGAAGCAAGCCGGC----UGG ((...--..))..((......(((.((((((((((((((..........)))))))).)))----))))))..((((((((........)))))))----))) ( -33.50) >DroYak_CAF1 3679 100 + 1 GCUC---CUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCAAAAGGCAGGCAGGCAGUCAAGUCGGCUAAGAAGCAAGCCGGCUGGCUGG ...(---((((((((...(((((..(((.(((.........)))..)))..))))).)).)))))))((((((.(((((((........))))))))))))). ( -42.10) >consensus GCUCCUCCUGCUUCCGUAUUUGCUCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA____GGCAGUCAAGUCGGCUAAGAAGCAAGCCGGC____UGG ((.......))..(((.....(((.((.(((((((..(((..((((((.((((.............))))))))))..))).))))))).)).)))....))) (-29.58 = -29.34 + -0.24)

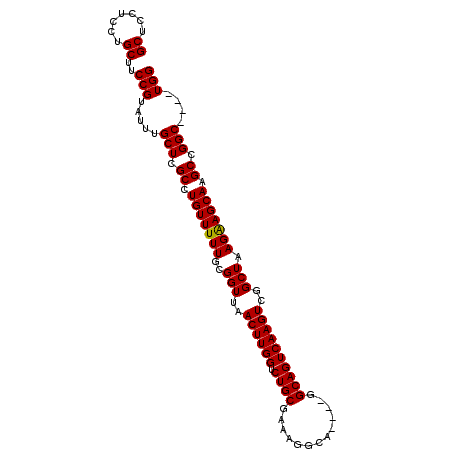
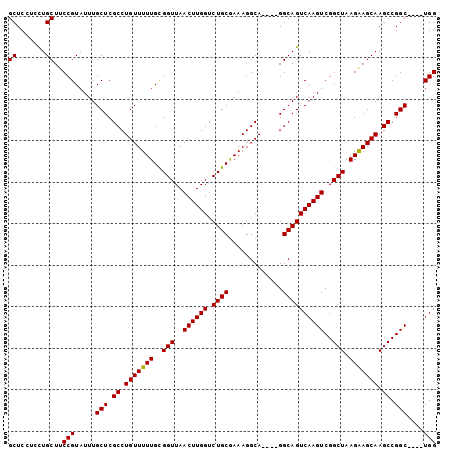
| Location | 11,566,827 – 11,566,937 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11566827 110 + 20766785 UCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGGAGCAAGCCGGC----UGGCAUUGAAGUUAGCAUAGAACACAAACAAGCUGGCGGCG (((((((((((....((((((((.((((((.....)))----))).((((.(((((((........)))))))----))))....))))))))..))))))..........))))).. ( -42.60) >DroPse_CAF1 3292 99 + 1 CCGCCUGUUUUUGUGGUAAAGUUGGUCUGGGAU---------CCCAGUCGAGUCGAG------UCGAGCCGGC----UUGCAUUGAAGUUGGCAUAAAAGACAAACAAGCUUGCGGCG ((((..((..((((.......((((.((((...---------.))))))))(((...------....((((((----((......))))))))......)))..))))))..)))).. ( -30.72) >DroSim_CAF1 3642 110 + 1 UCGCCUGUUUUUGUGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGGAGCAAGCCGGC----UGGCAUUGAAGUUAGCAUAGAACACAAACAAGCUGGCGGCG (((((.(((((((((((((((((.((((((.....)))----))).((((.(((((((........)))))))----))))....))))))))......)))))..)))).))))).. ( -45.50) >DroEre_CAF1 3626 110 + 1 UCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA----GGCAGUCAAGUCGGCUAAGAAGCAAGCCGGC----UGGCAUUGAAGUUAGCAUAGCACACAAACAAGCUGGCGGCG ((((((((((.(((.((((((((.((((((.....)))----))).((((.(((((((........)))))))----))))....))))))))...)))...)))))....))))).. ( -45.70) >DroYak_CAF1 3699 118 + 1 UCGCCUGUUUUUGCGGUUAACUUGGUCUGCAAAAGGCAGGCAGGCAGUCAAGUCGGCUAAGAAGCAAGCCGGCUGGCUGGCAUUGAAGUUAGCAUAGAACACAAACAAGCUGGCGGCG (((((((((((....((((((((.((((((.....)))))).(.((((((.(((((((........))))))))))))).)....))))))))..))))))..........))))).. ( -48.70) >DroPer_CAF1 3266 99 + 1 CCGCCUGUUUUUGUGGUAAAGUUGGUCUGGGAU---------CCCAGUCGAGUCGAG------UCGAGCCGGC----UUGCAUUGAAGUUGGCAUAAAAGACAAACAAGCUUGCGGCG ((((..((..((((.......((((.((((...---------.))))))))(((...------....((((((----((......))))))))......)))..))))))..)))).. ( -30.72) >consensus UCGCCUGUUUUUGCGGUUAACUUGGUCUGCGAAAGGCA____GGCAGUCAAGUCGGCUAAG_AGCAAGCCGGC____UGGCAUUGAAGUUAGCAUAGAACACAAACAAGCUGGCGGCG .((((..((..(((.......((((.((((.............))))))))(((((((........)))))))......)))..)).((((((...............)))))))))) (-27.68 = -27.88 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:59 2006