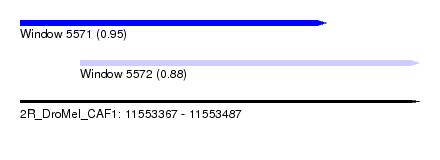
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,553,367 – 11,553,487 |
| Length | 120 |
| Max. P | 0.945965 |

| Location | 11,553,367 – 11,553,459 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.81 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -12.48 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
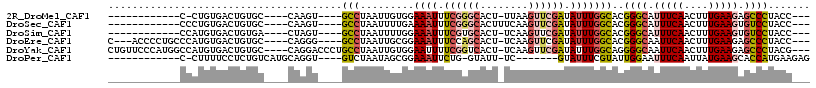
>2R_DroMel_CAF1 11553367 92 + 20766785 ------------C-CUGUGACUGUGC----CAAGU----GCCUAAUUGUGGAAAUUUCGGGCACU-UUAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGAGCCCUACC--- ------------.-........((((----(((((----(((.......)).....((((((...-....))))))))))))))))((((.(((((....))))).))))....--- ( -31.40) >DroSec_CAF1 13637 94 + 1 ------------CCCUGUGACUGUGC----CAAGU----GCCUAAUUUUGAAAAUUUCGGGCACUUUCAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGUGUCCUACC--- ------------..........((((----(((((----(((..(...(....)..)..)))))..((......))...)))))))((((((((((....))))))))))....--- ( -31.90) >DroSim_CAF1 13667 93 + 1 ------------CCAUGUGACUGUGA----CUAGU----GCCUAAUUUUGGAAAUUUCGUGCACU-UCAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGUGUCCUACC--- ------------.(((......))).----...((----(((.(((.(((((......(....).-.....))))).))).)))))((((((((((....))))))))))....--- ( -25.00) >DroEre_CAF1 13737 102 + 1 C---ACCCCUGCCCAUGUGACUGUGC----CAGGG----GCCUAAUUGCGGAAAUUUCCAGCACU-UCAAGUUCGAUAUUUGGCACGGGCAAUUCAACUUUGAAGAGCCCUACC--- .---.((((((..(((......))).----)))))----)......((((((....)))......-.(((((.....)))))))).((((..((((....))))..))))....--- ( -28.90) >DroYak_CAF1 13738 109 + 1 CUGUUCCCAUGGCCAUGUGACUGUGC----CAGGACCCUGCCUAAUUGUGGAAUUUUCGGUCACU-UCAAGUUCGAUAUUUGGCAGGGGCAAUUCAACUUUGAAGAGCCCUACG--- ......((.((((((......)).))----))))...(((((.((.(((.((((((..(....).-..)))))).))).)))))))((((..((((....))))..))))....--- ( -32.70) >DroPer_CAF1 13276 91 + 1 ------------C-CUUUUCCUCUGUCAUGCAGGU----GUCUAAUAGCGGAAAUUCUG-GUAUU-UC-------GUAUUUCGUAUUGGAAUUUCAAUUAUGAAGCACCAUGAAGAG ------------.-......((((.((((...(((----((...((((..(((((((..-((((.-..-------.......))))..)))))))..))))...))))))))))))) ( -25.70) >consensus ____________CCAUGUGACUGUGC____CAAGU____GCCUAAUUGUGGAAAUUUCGGGCACU_UCAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGAGCCCUACC___ .......................................(((.........((((.((((((........)))))).)))))))..((((.(((((....))))).))))....... (-12.48 = -13.38 + 0.91)

| Location | 11,553,385 – 11,553,487 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11553385 102 + 20766785 GCCUAAUUGUGGAAAUUUCGGGCACU-UUAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGAGCCCUACCUA-----UCCCGGCCCAGCAGCCACCACCACCC ........((((((((.((((((...-....)))))).))))(((..((((.(((((....))))).))))..((..-----....)).......))).))))..... ( -29.30) >DroSec_CAF1 13656 89 + 1 GCCUAAUUUUGAAAAUUUCGGGCACUUUCAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGUGUCCUACCUA-----UCCCGGCCCA--------------CC (((.........((((.((((((........)))))).))))((...((((((((((....))))))))))..))..-----....)))...--------------.. ( -25.20) >DroSim_CAF1 13686 87 + 1 GCCUAAUUUUGGAAAUUUCGUGCACU-UCAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGUGUCCUACCUA-----UCCCGACCCA--------------C- ........((((.......((((...-.(((((.....)))))))))((((((((((....))))))))))......-----..))))....--------------.- ( -22.30) >DroEre_CAF1 13765 90 + 1 GCCUAAUUGCGGAAAUUUCCAGCACU-UCAAGUUCGAUAUUUGGCACGGGCAAUUCAACUUUGAAGAGCCCUACCCCACUUUACCGCCCCC----------------- ........((((((((.((.(((...-....))).)).))))((...((((..((((....))))..))))....))......))))....----------------- ( -20.50) >DroYak_CAF1 13773 93 + 1 GCCUAAUUGUGGAAUUUUCGGUCACU-UCAAGUUCGAUAUUUGGCAGGGGCAAUUCAACUUUGAAGAGCCCUACGCUACCUUGCCGCACCCA--------------CC (((.((.(((.((((((..(....).-..)))))).))).))))).(((((..((((....))))..)))))..((......))........--------------.. ( -22.80) >consensus GCCUAAUUGUGGAAAUUUCGGGCACU_UCAAGUUCGAUAUUUGGCACGGGCAUUUCAACUUUGAAGAGCCCUACCUA_____UCCCGACCCA______________CC (((.........((((.((((((........)))))).)))))))..((((.(((((....))))).))))..................................... (-16.42 = -17.26 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:54 2006