| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,551,046 – 11,551,158 |
| Length | 112 |
| Max. P | 0.820175 |

| Location | 11,551,046 – 11,551,158 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
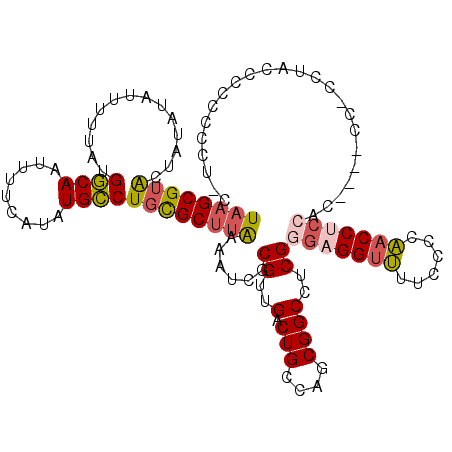
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.28 |
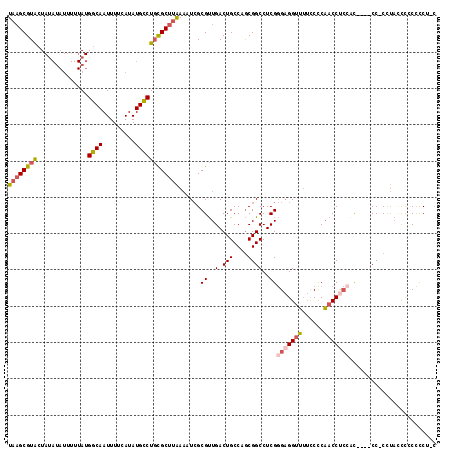
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
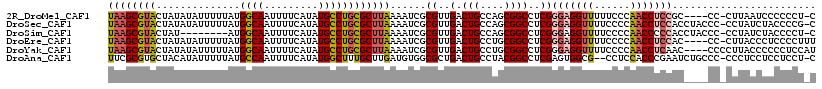
>2R_DroMel_CAF1 11551046 112 - 20766785 UAAGCGUACUAUAUAUUUUUAUGGCAAUUUUCAUAUGCCUGCGCUUAAAAUCGCGUUGACUGCCAGCGGCCUCGGGAGGUUUUUCCCAACCUCCGC----CC-CUUAAUCCCCCCU-C ((((((((..(((......)))((((.........)))))))))))).......((((.....))))(((....(((((((......)))))))))----).-.............-. ( -28.00) >DroSec_CAF1 11254 116 - 1 UAAGCGUACUAUAUAUUUUUAUGGCAAUUUUCAUAUGCCUGCGCUUAAAAUCGCGUUGACUGCCAGCGGCCUCGGGAGGUUUUCCCCAACCUCCACCUACCC-CCUAUCUACCCCG-C ((((((((..(((......)))((((.........)))))))))))).....((((((.....))))((.....(((((((......))))))).....)).-............)-) ( -27.90) >DroSim_CAF1 11325 108 - 1 UAAGCGUACUAU--------AUGGCAAUUUUCAUAUGCCUGCGCUUAAAAUCGCGUUGACUGCCAGCGGCCUCGGGAGGUUUUCCCCAACCCCCACCUACCC-CCUAUCUACCCCU-C ((((((((.(((--------((((......)))))))..))))))))......(((((.....))))).....(((.((((......)))))))........-.............-. ( -27.00) >DroEre_CAF1 11298 113 - 1 UAAGCGUACUAUAUAUUUUUAUGGCAAUUUUCAUAUGCCUGCGCUUAAAAUCGCGUUGACUGCCUGCGGCCUCGGGAGGUUUUCCCCAACCUCCAC----CC-CUUACCCUCCCCUUU ((((((((..(((......)))((((.........))))))))))))......((..(.(((....))))..))(((((((......)))))))..----..-............... ( -26.00) >DroYak_CAF1 11351 114 - 1 UAAGCGUACUAUAUAUUUUUAUGGCAAUUUUCAUAUGCCUGCGCUUAAAAUCGCGUUGACUGCCUGCGGCCUCGGGAGGUUUUCCCCAACCUCAAC----CCCCUUACCCCCCUCCAU ((((((((..(((......)))((((.........))))))))))))......((..(.(((....))))..)).((((((......))))))...----.................. ( -22.70) >DroAna_CAF1 9602 114 - 1 UUCGCGUGCUACAUAUUUUUAUGCCAAUUUUCAUAUGGCUUUGCUUGAUGUGGCGCUGACUGCCUACGGCCUCGAGUGGCG--CCUCCACCCGAAUCUGCCC-CCCUCCUCCUCCU-C .(((.(((((((((........((((.........))))........))))))))))))........(((.(((.((((..--...)))).)))....))).-.............-. ( -29.69) >consensus UAAGCGUACUAUAUAUUUUUAUGGCAAUUUUCAUAUGCCUGCGCUUAAAAUCGCGUUGACUGCCAGCGGCCUCGGGAGGUUUUCCCCAACCUCCAC____CC_CCUACCCCCCCCU_C ((((((((..............((((.........))))))))))))......((..(.(((....))))..))(((((((......)))))))........................ (-18.75 = -19.28 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:51 2006