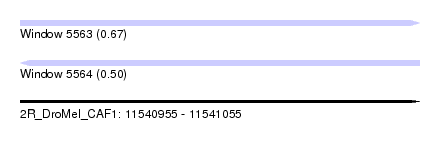
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,540,955 – 11,541,055 |
| Length | 100 |
| Max. P | 0.666814 |

| Location | 11,540,955 – 11,541,055 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.62 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
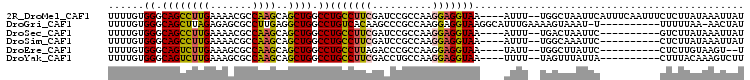
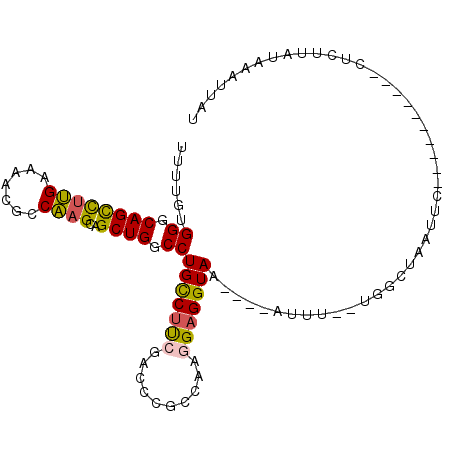
>2R_DroMel_CAF1 11540955 100 + 20766785 AUAAUUUAUAAGAGAAAUUGAAAUGAAUUAGCCA--AAAU----UUACCUCCUUGGCGGAUCGAAGGCAGGCCAGCUGCUUGGCGUUUUCAAGGCUGCCCACAAAA .((((((((..((....))...))))))))(((.--..((----(..((.....))..)))....))).((.((((..(((((.....))))))))).))...... ( -23.40) >DroGri_CAF1 1156 94 + 1 AUAGUU-UUAAAAA----------A-AUUUACUUUUCAAAUGCCUUACCUCCUUGGCGGGCUUGUGACAGGCCAGCCUCAAGGCGCUCUCUAAGCUGCCCACAAAA ......-.......----------.-...............((((..((.....)).))))(((((...(((..(((....)))(((.....))).)))))))).. ( -20.10) >DroSec_CAF1 1122 90 + 1 AUAAUUUAUAAGAC----------GAAUUAGUCA--AAAU----UUACCUCCUUGGCGGAUCGAAGGCAGGCCAGCUGCUUGGCGUUUUCAAGGCUGCCCACAAAA ...........(((----------......))).--....----..........(((((...((((((..(((((....)))))))))))....)))))....... ( -20.80) >DroSim_CAF1 1120 90 + 1 AUAAUUUAUAAGAG----------GAAUUUGCCA--AAAU----UUACCUCCUUGGCGGAUCGAAGGCAGGCCAGCUGCUUGGCGUUUUCAAGGCUGCCCACAAAA ...........(((----------((((((....--))))----)..))))...(((((...((((((..(((((....)))))))))))....)))))....... ( -22.60) >DroEre_CAF1 1120 88 + 1 A--ACUUACAAGAG----------GAAUAAGCCA--AAUA----UUACCUCCUUGGCGGGUCUAAGGCAGGCCAGCUGCUUGGCGCUUUCAAGACUGCCCACAAAA .--........(((----------(((((.....--..))----)).))))...(((((.((((((((..(((((....))))))))))..))))))))....... ( -23.40) >DroYak_CAF1 1121 90 + 1 AAGACUUUGUAAAG----------UAAUAAACUA--AAAA----UUACCUCCUUGGCAGGUCGAAGGCAGGCCAGCUGCUUGGCGCUUUCAAGACUGCCCACAAAA .....(((((...(----------((((......--...)----))))......(((((...((((((..(((((....)))))))))))....))))).))))). ( -23.70) >consensus AUAAUUUAUAAGAG__________GAAUUAGCCA__AAAA____UUACCUCCUUGGCGGAUCGAAGGCAGGCCAGCUGCUUGGCGCUUUCAAGGCUGCCCACAAAA ......................................................(((((.((((((((..(((((....)))))))))))..)))))))....... (-15.70 = -15.62 + -0.08)


| Location | 11,540,955 – 11,541,055 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11540955 100 - 20766785 UUUUGUGGGCAGCCUUGAAAACGCCAAGCAGCUGGCCUGCCUUCGAUCCGCCAAGGAGGUAA----AUUU--UGGCUAAUUCAUUUCAAUUUCUCUUAUAAAUUAU .((((((((.((..((((((..((......))((((((((((((..........))))))).----....--.))))).....))))))...)))))))))).... ( -22.20) >DroGri_CAF1 1156 94 - 1 UUUUGUGGGCAGCUUAGAGAGCGCCUUGAGGCUGGCCUGUCACAAGCCCGCCAAGGAGGUAAGGCAUUUGAAAAGUAAAU-U----------UUUUUAA-AACUAU ..(((((((((((((.(((.....))).))))).)))...)))))(((.(((.....)))..))).(((((((((.....-)----------)))))))-)..... ( -26.10) >DroSec_CAF1 1122 90 - 1 UUUUGUGGGCAGCCUUGAAAACGCCAAGCAGCUGGCCUGCCUUCGAUCCGCCAAGGAGGUAA----AUUU--UGACUAAUUC----------GUCUUAUAAAUUAU ......((.((((((((.......))))..)))).))(((((((..........)))))))(----((((--.(((......----------)))....))))).. ( -22.20) >DroSim_CAF1 1120 90 - 1 UUUUGUGGGCAGCCUUGAAAACGCCAAGCAGCUGGCCUGCCUUCGAUCCGCCAAGGAGGUAA----AUUU--UGGCAAAUUC----------CUCUUAUAAAUUAU .(((((((.((((((((.......))))..)))).))(((((((..........))))))).----....--..)))))...----------.............. ( -22.60) >DroEre_CAF1 1120 88 - 1 UUUUGUGGGCAGUCUUGAAAGCGCCAAGCAGCUGGCCUGCCUUAGACCCGCCAAGGAGGUAA----UAUU--UGGCUUAUUC----------CUCUUGUAAGU--U .......(((.((((....((.((((......)))))).....))))..))).((((((.((----((..--.....)))))----------)))))......--. ( -24.50) >DroYak_CAF1 1121 90 - 1 UUUUGUGGGCAGUCUUGAAAGCGCCAAGCAGCUGGCCUGCCUUCGACCUGCCAAGGAGGUAA----UUUU--UAGUUUAUUA----------CUUUACAAAGUCUU ((((((.(((((..(((((.((((((......))))..)).))))).)))))...(((((((----(...--......))))----------)))))))))).... ( -26.40) >consensus UUUUGUGGGCAGCCUUGAAAACGCCAAGCAGCUGGCCUGCCUUCGACCCGCCAAGGAGGUAA____AUUU__UGGCUAAUUC__________CUCUUAUAAAUUAU ......((.((((((((.......))))..)))).))(((((((..........)))))))............................................. (-16.36 = -16.12 + -0.25)
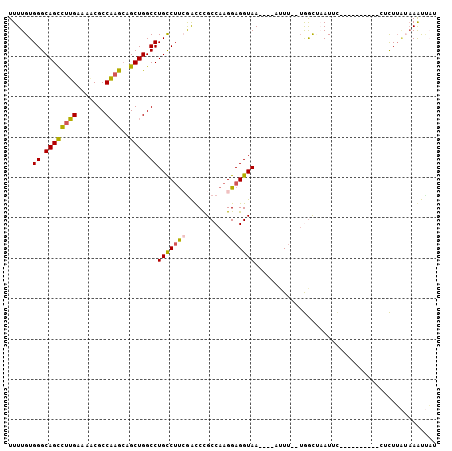
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:46 2006