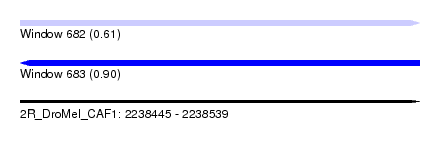
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,238,445 – 2,238,539 |
| Length | 94 |
| Max. P | 0.903031 |

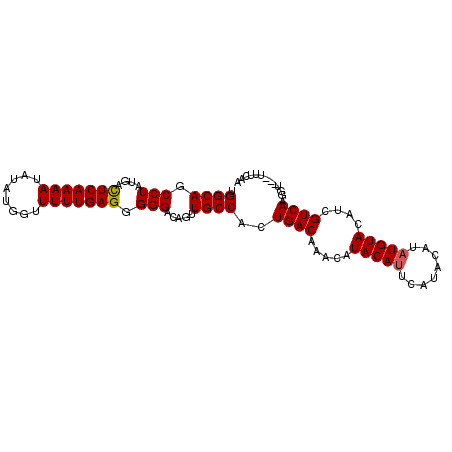
| Location | 2,238,445 – 2,238,539 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2238445 94 + 20766785 UGGCAGCCUAUGAUUCAAAAUAUAUGGUUUUUGAGGGGGACAUAUGCCUCUGACGAACAUACA----UACAUAUGUACAUCGUCAAGGAUAUAUUCAA .((((.(((....(((((((........))))))).))).....))))..((((((...((((----(....)))))..))))))............. ( -23.50) >DroSec_CAF1 4999 95 + 1 UGGCAGCCUAUGACUCAAAAUAUAUGGUUUUUGAGGGGGACAGUUGCCACUGACAAACAUACAUUCAUAAAUAUGUACAUCGUCAAGGU---UUUCAA (((((((......(((((((........)))))))(....).))))))).((((.....(((((........)))))....))))....---...... ( -23.90) >DroSim_CAF1 5010 95 + 1 UGGCAGCCUAUGACUCAAAAUAUAUGGUUUUUGAGGGGGUCAGUUGCCACUGACAAACAUACAUUCAUACAUAUGUACAUCGUCAAGGU---UUUCAA (((.(((((.((((.....(((((((.....((((.(.((((((....))))))...).....))))..))))))).....))))))))---).))). ( -24.30) >consensus UGGCAGCCUAUGACUCAAAAUAUAUGGUUUUUGAGGGGGACAGUUGCCACUGACAAACAUACAUUCAUACAUAUGUACAUCGUCAAGGU___UUUCAA .((((.(((....(((((((........))))))).))).....))))..((((.....(((((........)))))....))))............. (-19.05 = -19.17 + 0.11)

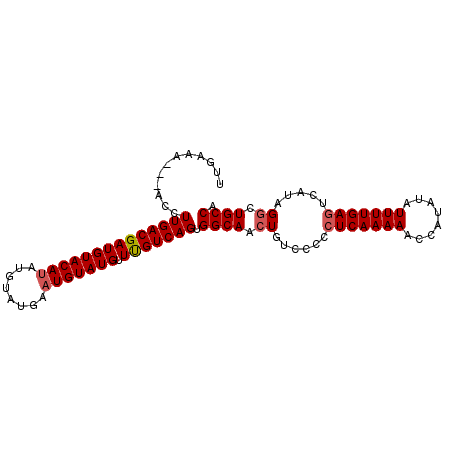
| Location | 2,238,445 – 2,238,539 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2238445 94 - 20766785 UUGAAUAUAUCCUUGACGAUGUACAUAUGUA----UGUAUGUUCGUCAGAGGCAUAUGUCCCCCUCAAAAACCAUAUAUUUUGAAUCAUAGGCUGCCA .(((((((((((((((((((((((((....)----)))))).)))))).))).)))))).....((((((........)))))).))).......... ( -21.80) >DroSec_CAF1 4999 95 - 1 UUGAAA---ACCUUGACGAUGUACAUAUUUAUGAAUGUAUGUUUGUCAGUGGCAACUGUCCCCCUCAAAAACCAUAUAUUUUGAGUCAUAGGCUGCCA ......---...((((((((((((((........))))))).)))))))(((((.((((....(((((((........)))))))..))))..))))) ( -24.50) >DroSim_CAF1 5010 95 - 1 UUGAAA---ACCUUGACGAUGUACAUAUGUAUGAAUGUAUGUUUGUCAGUGGCAACUGACCCCCUCAAAAACCAUAUAUUUUGAGUCAUAGGCUGCCA ......---...((((((((((((((........))))))).)))))))(((((.((......(((((((........))))))).....)).))))) ( -23.40) >consensus UUGAAA___ACCUUGACGAUGUACAUAUGUAUGAAUGUAUGUUUGUCAGUGGCAACUGUCCCCCUCAAAAACCAUAUAUUUUGAGUCAUAGGCUGCCA ............((((((((((((((........))))))).))))))).((((.((......(((((((........))))))).....)).)))). (-20.92 = -21.70 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:56 2006