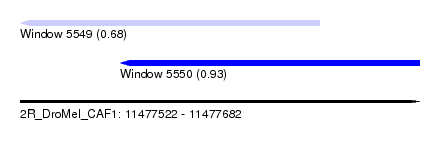
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,477,522 – 11,477,682 |
| Length | 160 |
| Max. P | 0.934129 |

| Location | 11,477,522 – 11,477,642 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
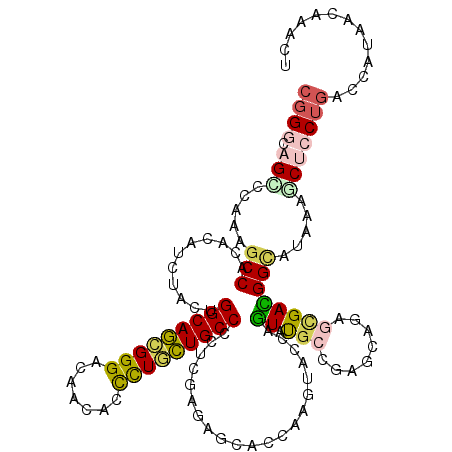
>2R_DroMel_CAF1 11477522 120 - 20766785 CUGGCAGUCCAAAGCCACAUAUUUACUGGCAGAGGGAUAAUACCCUACUGCCCAUCGAGGGCACCAAGUACCAGUAUGAGGAGCAAAGCGACGGAAUAAAACUCCUAACCAUUAACAACU .((((........))))((((((((((((.((((((......)))).))((((.....)))).)).))))..))))))(((((..................))))).............. ( -30.27) >DroVir_CAF1 33816 120 - 1 CGGGCAGCCCCAAGCCGCAUAUCUCCUGGCAGCGGGACAAUACCCUGCUGCCCCUCGAAACAGCUAAGUACGAGUACGCGGAGCACAGCGAUGGCAUCAAGCAGCUGAGCAUCACAUCCU .(((....)))...((((.........(((((((((.......))))))))).((((..((......)).))))...)))).((.((((....((.....)).)))).)).......... ( -42.60) >DroPse_CAF1 17257 120 - 1 CGGGAAGCCCAAAGCCACACAUCUACUGGCAGCGGGACAACACCUUGCUGCCGCUGGAGAGCAAAAAGUACCAGUAUGCCGAGCAGAGUGACGGCGUAAAGCUCCUGACCAUAGCGAACU (((((.((.....(((.(((.(((.(((((((((((.......)))))))))(((((...((.....)).)))))......)).))))))..))).....)))))))............. ( -40.30) >DroGri_CAF1 27999 120 - 1 CGGGCAGCCCCAAGCCAAACAUCUACUGGCAACGGGAUAAUAGCCUGUUGCCCGUCGAUACCGCCAAGUAUCAAUAUGCCGAGCAAAGCGAUGGUGUCAAACAGCUGACCAUAACAUCCU .((.((((................((.(((((((((.......))))))))).)).(((((((.(..((((....))))...((...))).))))))).....)))).)).......... ( -34.20) >DroAna_CAF1 18192 120 - 1 CGGGCAGUCCCAAGCCCAAAAUCUACUGGCAGAGGGAUAACACUCUUCUGCCGCUGGAGAGUCCCAAGUACCAGUUCUCCGAGCAGGCUGACGGCACGAAGCUCCUGACCAUUGCCCAGU .((((((((...((((...........(((((((((.......)))))))))(((((((((.............)))))).))).)))))))((((.(.....).)).))..)))))... ( -44.42) >DroPer_CAF1 17268 120 - 1 CGGGAAGCCCAAAGCCACACAUCUACUGGCAGCGGGACAACACCUUGCUGCCGCUGGAGAGCAAAAAGUACCAGUAUGCCGAGCAGAGUGACGGCGUAAAUCUCCUGACCAUAGCGAACU (((((........(((.(((.(((.(((((((((((.......)))))))))(((((...((.....)).)))))......)).))))))..))).......)))))............. ( -36.96) >consensus CGGGCAGCCCAAAGCCACACAUCUACUGGCAGCGGGACAACACCCUGCUGCCCCUCGAGAGCACCAAGUACCAGUAUGCCGAGCAGAGCGACGGCAUAAAGCUCCUGACCAUAACAAACU ((((.(((.....(((...........(((((((((.......))))))))).....................((.(((........)))))))).....)))))))............. (-21.39 = -21.65 + 0.26)

| Location | 11,477,562 – 11,477,682 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
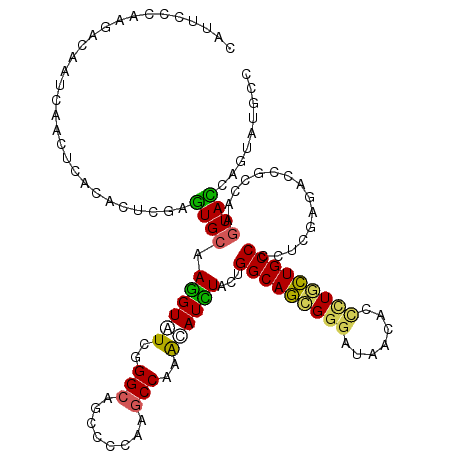
>2R_DroMel_CAF1 11477562 120 - 20766785 CAUUCCCAGGGUAACCAGCUCACAUUGGAGUGCAAGGUAUCUGGCAGUCCAAAGCCACAUAUUUACUGGCAGAGGGAUAAUACCCUACUGCCCAUCGAGGGCACCAAGUACCAGUAUGAG ........((....))..((((.(((((.((((.((((((.((((........)))).))))))...(((((((((......)))).)))))........))))......))))).)))) ( -37.70) >DroVir_CAF1 33856 120 - 1 CAUUCCCAAGAAAAUCGACUUGUACUCGAGUGCAAGGUCUCGGGCAGCCCCAAGCCGCAUAUCUCCUGGCAGCGGGACAAUACCCUGCUGCCCCUCGAAACAGCUAAGUACGAGUACGCG ...............((((((((((((((((((..((.((.(((....))).)))))))........(((((((((.......))))))))).))))).........)))))))).)).. ( -40.60) >DroGri_CAF1 28039 120 - 1 CAUUCCCAAGACAAUCAAUUAACACUUGAAUGCAAGGUCUCGGGCAGCCCCAAGCCAAACAUCUACUGGCAACGGGAUAAUAGCCUGUUGCCCGUCGAUACCGCCAAGUAUCAAUAUGCC .......................(((((...((..((.((.(((....))).))))....(((.((.(((((((((.......))))))))).)).)))...)))))))........... ( -31.70) >DroMoj_CAF1 26660 120 - 1 CAUUCCCAAGACAAUCGCCUUACGCUCGAGUGCAAAGUGUCCGGCAGUCCCAAGCCAAGCAUUUACUGGCAGCGAGAUAACACACUGCUGCCCCUGGAUACGGCCAAAUAUGAAUAUGCC ................(((....((......))...(((((((((........)))...........((((((.((........))))))))...)))))))))................ ( -31.60) >DroAna_CAF1 18232 120 - 1 CAUUCCCAGGCCAAUCAGCUGGCUUUAGAGUGCAAGGUUUCGGGCAGUCCCAAGCCCAAAAUCUACUGGCAGAGGGAUAACACUCUUCUGCCGCUGGAGAGUCCCAAGUACCAGUUCUCC (((((..((((((......))))))..)))))..((((((.((((........)))).))))))...(((((((((.......)))))))))...((((((.............)))))) ( -41.92) >DroPer_CAF1 17308 120 - 1 CAUUCACAGGGCAAUAGCCUCACGCUGGAGUGCAAGGUAUCGGGAAGCCCAAAGCCACACAUCUACUGGCAGCGGGACAACACCUUGCUGCCGCUGGAGAGCAAAAAGUACCAGUAUGCC ........((((....))))((..((((.(((...(((...(((...)))...))).)))...(((((((((((((.......)))))))))(((....)))....))))))))..)).. ( -40.30) >consensus CAUUCCCAAGACAAUCAACUCACACUCGAGUGCAAGGUAUCGGGCAGCCCCAAGCCAAACAUCUACUGGCAGCGGGAUAACACCCUGCUGCCCCUCGAGACCGCCAAGUACCAGUAUGCC .............................((((.((((((..(((........)))..))))))...(((((((((.......)))))))))...............))))......... (-23.41 = -23.00 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:32 2006