| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,460,098 – 11,460,192 |
| Length | 94 |
| Max. P | 0.859349 |

| Location | 11,460,098 – 11,460,192 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -18.48 |
| Energy contribution | -19.62 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
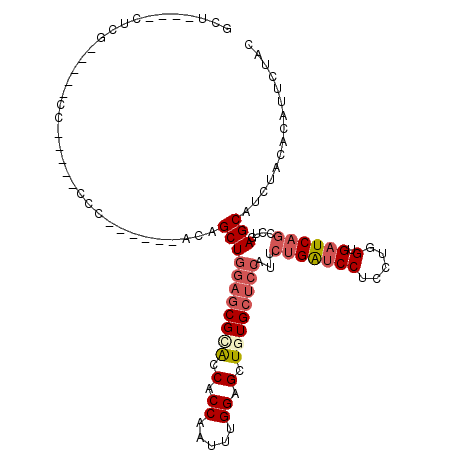
>2R_DroMel_CAF1 11460098 94 + 20766785 GCU----UUCG-----CC-----CCC------ACAGCUGGAGCGCACCACCAAUUUGGAGCUAUGCUCCAUUCUGAUCCUCCUGGUGAUCAGCCUGAGCAUCUACACAUUCUAC (((----(..(-----(.-----...------...))..))))((.(((......))).)).((((((....(((((((.....).))))))...))))))............. ( -25.40) >DroVir_CAF1 13869 103 + 1 ACC----CUCU-AUCUCCCCUCACAU------AUAGCUGGAACGUGCCACCAAAUCGGAGCUGUGCAGCGCUUUGGCCCUGUUGGUGCUCUCGCUGAGCAUCUAUGCCUUCUAC ...----....-..............------...((((..(((.((..((.....)).))))).)))).....(((......(((((((.....)))))))...)))...... ( -24.30) >DroSec_CAF1 1734 94 + 1 GCU----UUCG-----UC-----CCC------ACAGCUGGAGCGCACCACCAAUUUGGAGCUGUGCUCCAUUCUGAUCCUCCUGGUGAUCAGCCUGAGCAUCUACACAUUCUAC (((----(...-----..-----...------.....(((((((((.(.((.....)).).)))))))))..(((((((.....).))))))...))))............... ( -27.50) >DroSim_CAF1 1109 94 + 1 GCU----UUCG-----UC-----CCC------ACAGCUGGAGCGCACCACCAAUUUGGAGCUGUGCUCCAUUCUGAUCCUCCUGGUGAUCAGCCUGAGCAUCUACACAUUCUAC (((----(...-----..-----...------.....(((((((((.(.((.....)).).)))))))))..(((((((.....).))))))...))))............... ( -27.50) >DroEre_CAF1 3213 98 + 1 UCCUACCCUCC-----CC-----CCA------ACAGCUGGAGCGGACCACCAAGUUGGAGCUGUGCUCCAUCCUGAUCCUCCUGGUGAUCAGCCUGAGCAUCUACACAUUCUAC ...........-----..-----(((------((.((....))((....))..)))))....((((((....(((((((.....).))))))...))))))............. ( -23.70) >DroYak_CAF1 1154 108 + 1 UCUUACCCCCCUUCC-UC-----CCACCCAUCACAGCUGGAGCGCACCACCAAGUCGGAGCUGUGCUCGAUCCUGAUCCUGCUGGUGAUCAGCCUGAGCAUCUACACAUUCUAC ...............-..-----.........((((((((......)).((.....))))))))(((((...(((((((.....).))))))..)))))............... ( -23.70) >consensus GCU____CUCG_____CC_____CCC______ACAGCUGGAGCGCACCACCAAUUUGGAGCUGUGCUCCAUUCUGAUCCUCCUGGUGAUCAGCCUGAGCAUCUACACAUUCUAC ...................................(((((((((((.(.((.....)).).))))))))...(((((((.....).))))))....)))............... (-18.48 = -19.62 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:29 2006