| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,449,567 – 11,449,661 |
| Length | 94 |
| Max. P | 0.775985 |

| Location | 11,449,567 – 11,449,661 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
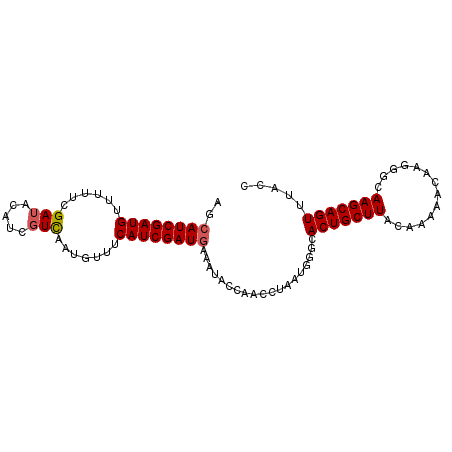
>2R_DroMel_CAF1 11449567 94 + 20766785 AGCAUCGAUGUUUUUCGAUACAUCGUCAAUGUUUCAUCGAUGAAAUACCAACUUAAUGGGCACUGCUUACAAAAACAAGGUCAAGCAGUUUACC .((..((((((........))))))....(((((((....)))))))(((......)))))(((((((((.........)).)))))))..... ( -21.30) >DroSec_CAF1 1972 94 + 1 AGCAUCGAUGAUUUUCGAUACAUCGUCAAUGUUUCAUCGAUAAAAUACCAACCUAAUGGGCACUGCUUACAAAAACCAAGGCAAGCAGUUUACG .((((((((((..(.(((....))).....)..))))))))......(((......)))))(((((((.(..........).)))))))..... ( -21.00) >DroSim_CAF1 1970 94 + 1 AGCAUCGAUGAUUUUCGAUACAUCGUCAAUGUUUCAUCGAUGAAAUACCAACCUAAUGGGCACUGCUUACAAAAACAAAGGCAAGCAGUUUACG ..(((((((((..(.(((....))).....)..))))))))).....(((......)))..(((((((.(..........).)))))))..... ( -21.50) >DroEre_CAF1 2007 92 + 1 UACAUCGAUGUUUUUCGAUACA-CCUUAAUGUCUCAUCGAUGACACACCGUCUUAAUUUAAACUGCUUAGCGAAAGCAGUGCAAGCAGUU-AGC ..((((((((......((((..-......)))).))))))))..................((((((((.((....)).....))))))))-... ( -23.00) >DroYak_CAF1 1981 93 + 1 UGCAUCGAUGUUUUUCGAUAAA-CGUCAAUGUUUCAUCGAUGACAUACCGUCCUAAUGAAAACUGCUUGGCGAAAUUAGGGGAAGCAGUUCAGC .((((.(((((((......)))-)))).))))(((((.((((......))))...)))))((((((((..(........)..)))))))).... ( -22.40) >consensus AGCAUCGAUGUUUUUCGAUACAUCGUCAAUGUUUCAUCGAUGAAAUACCAACCUAAUGGGCACUGCUUACAAAAACAAGGGCAAGCAGUUUACC ..((((((((......(((.....))).......))))))))...................(((((((..............)))))))..... (-15.10 = -15.34 + 0.24)


| Location | 11,449,567 – 11,449,661 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
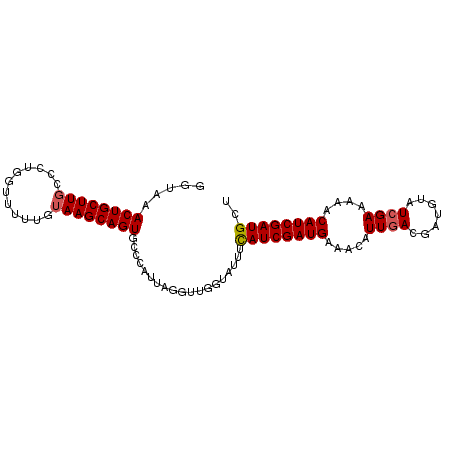
>2R_DroMel_CAF1 11449567 94 - 20766785 GGUAAACUGCUUGACCUUGUUUUUGUAAGCAGUGCCCAUUAAGUUGGUAUUUCAUCGAUGAAACAUUGACGAUGUAUCGAAAAACAUCGAUGCU ((...((((((((............))))))))..)).....((..((.(((((....))))).))..))...(((((((......))))))). ( -26.10) >DroSec_CAF1 1972 94 - 1 CGUAAACUGCUUGCCUUGGUUUUUGUAAGCAGUGCCCAUUAGGUUGGUAUUUUAUCGAUGAAACAUUGACGAUGUAUCGAAAAUCAUCGAUGCU .....(((((((((..........))))))))).........((..((.(((((....))))).))..))...(((((((......))))))). ( -24.30) >DroSim_CAF1 1970 94 - 1 CGUAAACUGCUUGCCUUUGUUUUUGUAAGCAGUGCCCAUUAGGUUGGUAUUUCAUCGAUGAAACAUUGACGAUGUAUCGAAAAUCAUCGAUGCU .....(((((((((..........))))))))).........((..((.(((((....))))).))..))...(((((((......))))))). ( -26.50) >DroEre_CAF1 2007 92 - 1 GCU-AACUGCUUGCACUGCUUUCGCUAAGCAGUUUAAAUUAAGACGGUGUGUCAUCGAUGAGACAUUAAGG-UGUAUCGAAAAACAUCGAUGUA ...-((((((((((.........)).))))))))..........((((((....((((((..((......)-).))))))...))))))..... ( -23.70) >DroYak_CAF1 1981 93 - 1 GCUGAACUGCUUCCCCUAAUUUCGCCAAGCAGUUUUCAUUAGGACGGUAUGUCAUCGAUGAAACAUUGACG-UUUAUCGAAAAACAUCGAUGCA ...(((((((((..(........)..)))))))))(((((..(((.....)))...)))))..((((((.(-(((......)))).)))))).. ( -21.00) >consensus GGUAAACUGCUUGCCCUGGUUUUUGUAAGCAGUGCCCAUUAGGUUGGUAUUUCAUCGAUGAAACAUUGACGAUGUAUCGAAAAACAUCGAUGCU .....((((((((............))))))))...................((((((((.....((((.......))))....)))))))).. (-16.62 = -16.86 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:22 2006