
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,449,004 – 11,449,110 |
| Length | 106 |
| Max. P | 0.891269 |

| Location | 11,449,004 – 11,449,110 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -23.31 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11449004 106 + 20766785 CAAUGA-CCCGCUAUGGUGAAUCACCA-GAAGCGA--------GCGACACCAGCUCAG-GGUACUCACCGGCCAGUCCUUGAAAGUUACCAAGCGAUUGGCCUCCAACAAGAGAUCG .....(-((((((.(((((...)))))-..)))((--------((.......)))).)-))).......((((((((((((........)))).))))))))............... ( -40.40) >DroGri_CAF1 1082 102 + 1 --------------UGCGGAAUGAGCAUGAAGCGUUUCCAUCUCCGUCAUAAACUCU-AUAUACGAACUGGCCAAUCCUUGAACGUUUCCAGGCGAUUCGCUUCGAUGAGCAGAUCC --------------...(((.((..(((((((((..(((.....(((.(((......-.))))))..((((..(((........))).))))).))..)))))).)))..))..))) ( -22.00) >DroSec_CAF1 1409 106 + 1 CAAAGA-CCCGCUUUGGUGAAUCACCA-GAAGCGA--------GCGACACCAGCUCAG-GGUACUCACCGGCCAGUCCUUGAAAGUUACCAAGCGAUUGGCCUCCAACAAGAGAUCG .....(-((((((((((((...)))))-).)))((--------((.......)))).)-))).......((((((((((((........)))).))))))))............... ( -41.00) >DroSim_CAF1 1407 106 + 1 CAAAGU-CCCGCUUUGGUGAAUCACCA-GAAGCGA--------GCGACACCAGCUCAG-GGUACUCACCGGCCAGUCCUUGAAAGUUACCAAGCGAUUGGCCUCCAACAAGAGAUCG ...(((-((((((((((((...)))))-).)))((--------((.......)))).)-)).)))....((((((((((((........)))).))))))))............... ( -41.00) >DroEre_CAF1 1443 107 + 1 CAAAGAGUCCGCUUUGGUGAAUCACCA-GAAGCAA--------GCGACACCAGGUCAG-AGUACUCACCGGCCAGUCCUUGAAGGUUUCCAAGCGAUUGGCCUCCAACAAGAGAUCG ....((((.((((((((((...)))))-).)))..--------..(((.....)))..-.).))))...((((((((((((........)))).))))))))............... ( -35.30) >DroYak_CAF1 1417 107 + 1 CAAAGAGCACGCUUUGGUGAAUCACCA-GAAGCAA--------GCGACACCAGCUCAG-GGUACUCACCGGCCAGUCCUUGAAGGUUUCCAAGCGAUUGGCCUCCAACAAGAGAUCG ....((((..(((((((((...)))))-).)))..--------))........(((..-(((....)))((((((((((((........)))).))))))))........))).)). ( -35.10) >consensus CAAAGA_CCCGCUUUGGUGAAUCACCA_GAAGCGA________GCGACACCAGCUCAG_GGUACUCACCGGCCAGUCCUUGAAAGUUACCAAGCGAUUGGCCUCCAACAAGAGAUCG ..........(((.(((((...)))))...)))....................(((...(((....)))((((((((((((........)))).))))))))........))).... (-23.31 = -24.45 + 1.14)

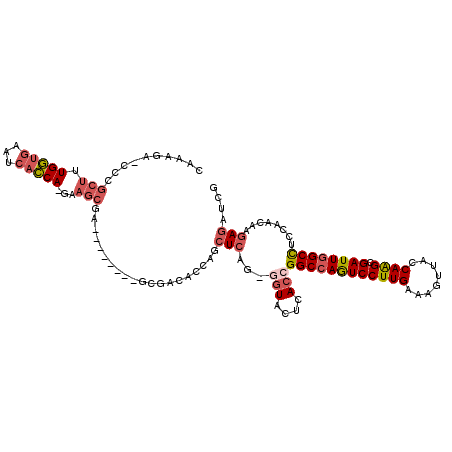
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:21 2006