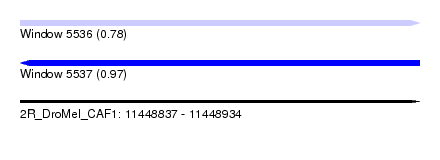
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,448,837 – 11,448,934 |
| Length | 97 |
| Max. P | 0.971569 |

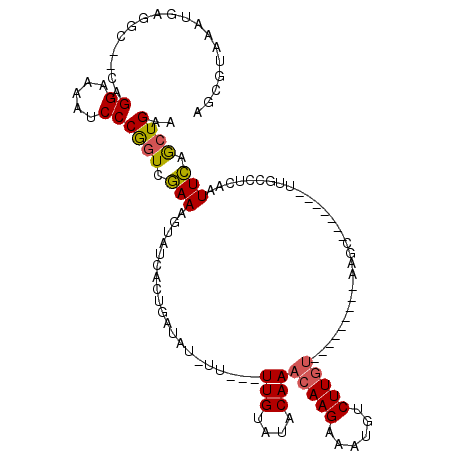
| Location | 11,448,837 – 11,448,934 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.91 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.22 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11448837 97 + 20766785 UUCAGCUGAAUUGAGGCA--------GCUA---------ACAAGACAUUUCUUGUUUGUAUACAA---AA-AUAUCAUUGAUACUUUCGACCGGGAUUUUCCUG--GCCUCAUUUACGCU ...((((((((.(((((.--------((.(---------((((((....))))))).))......---..-......((((.....)))).((((.....))))--)))))))))).))) ( -27.00) >DroSec_CAF1 1240 95 + 1 UUCAGCUGAAUUGAGGCAA-------GAC------------AAGAAAUUUCUUGUUUGUAUACAA---AA-AUAUCAGUGAUACUUUCGACCGGGAUUUCCCUG--GCCUCAUUUACGCU ...((((((((.(((((.(-------(((------------((((....))))))))((((((..---..-......)).))))........(((....)))..--)))))))))).))) ( -26.00) >DroSim_CAF1 1240 98 + 1 UUCAGCUAAAUUGAGGCAA-------GACA---------ACAAGAAAUUUCUUGUUUGUAUACAA---AA-AUAUCAGUGAUACUUUUGACCGGGAUUUUCCUG--GCGUCAUUUACGCU ...((((((((.((.((..-------.(((---------((((((....))))).))))...(((---((-.((((...)))).)))))..((((.....))))--)).))))))).))) ( -24.70) >DroEre_CAF1 1251 120 + 1 UUCAGCUGAAUUGAGUUAAUUAGGCAGCUUACAUAUUUCACAAGUCAUUUCUUGUUUGUAUACAAACAAACGUUUUAGAGAUUAUUUCUACUGGGUUUUCCCUUAUUUCUUAUUUACGCU ...(((((..((((.....)))).)))))......................(((((((....))))))).(((...((((((..........(((....)))..)))))).....))).. ( -18.50) >DroYak_CAF1 1226 115 + 1 UUCAGUUGAAUUGUGUUAAUUUGGCAGCUUACAUAUUUCACAAGACUUUUCUUGUUUGUAUACAA---AAGGUUUCCGAGAUUAUUUCGAAUGGGAUUUUCCUG--CGCUUAUUUACGCU ............((((.(((..(((.((....((((...((((((....))))))..))))....---.(((.(((((.((.....))...)))))....))))--)))).))).)))). ( -20.90) >consensus UUCAGCUGAAUUGAGGCAA_______GCUA_________ACAAGACAUUUCUUGUUUGUAUACAA___AA_AUAUCAGUGAUACUUUCGACCGGGAUUUUCCUG__GCCUCAUUUACGCU ...((((((((.(((((......................(((((......))))).....................................(((....)))....)))))))))).))) (-12.86 = -13.22 + 0.36)

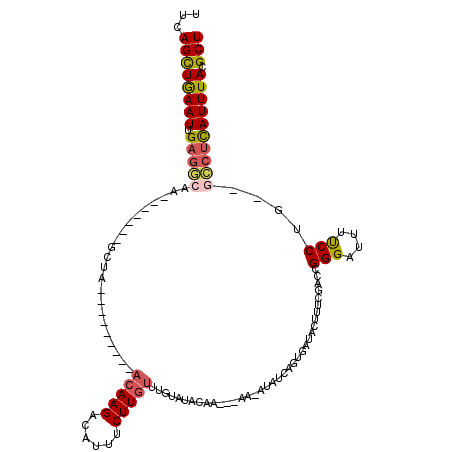
| Location | 11,448,837 – 11,448,934 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.91 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -7.94 |
| Energy contribution | -7.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11448837 97 - 20766785 AGCGUAAAUGAGGC--CAGGAAAAUCCCGGUCGAAAGUAUCAAUGAUAU-UU---UUGUAUACAAACAAGAAAUGUCUUGU---------UAGC--------UGCCUCAAUUCAGCUGAA (((.....((((((--..((......))(((((((((((((...)))))-))---)))......(((((((....))))))---------).))--------))))))).....)))... ( -26.90) >DroSec_CAF1 1240 95 - 1 AGCGUAAAUGAGGC--CAGGGAAAUCCCGGUCGAAAGUAUCACUGAUAU-UU---UUGUAUACAAACAAGAAAUUUCUU------------GUC-------UUGCCUCAAUUCAGCUGAA (((.....((((((--..(((....)))(..((((((((((...)))))-))---)))....)..((((((....))))------------)).-------..)))))).....)))... ( -28.40) >DroSim_CAF1 1240 98 - 1 AGCGUAAAUGACGC--CAGGAAAAUCCCGGUCAAAAGUAUCACUGAUAU-UU---UUGUAUACAAACAAGAAAUUUCUUGU---------UGUC-------UUGCCUCAAUUUAGCUGAA (((.(((((((.((--.((((.......(..((((((((((...)))))-))---)))....).(((((((....))))))---------).))-------)))).)).))))))))... ( -23.00) >DroEre_CAF1 1251 120 - 1 AGCGUAAAUAAGAAAUAAGGGAAAACCCAGUAGAAAUAAUCUCUAAAACGUUUGUUUGUAUACAAACAAGAAAUGACUUGUGAAAUAUGUAAGCUGCCUAAUUAACUCAAUUCAGCUGAA .(((((.(((((......(((....)))..((((.......)))).....((((((((....))))))))......)))))....))))).(((((...((((.....)))))))))... ( -21.00) >DroYak_CAF1 1226 115 - 1 AGCGUAAAUAAGCG--CAGGAAAAUCCCAUUCGAAAUAAUCUCGGAAACCUU---UUGUAUACAAACAAGAAAAGUCUUGUGAAAUAUGUAAGCUGCCAAAUUAACACAAUUCAACUGAA (((....(((..((--(((((........(((((.......)))))....((---((((......))))))....)))))))...)))....)))......................... ( -17.80) >consensus AGCGUAAAUGAGGC__CAGGAAAAUCCCGGUCGAAAGUAUCACUGAUAU_UU___UUGUAUACAAACAAGAAAUGUCUUGU_________AAGC_______UUGCCUCAAUUCAGCUGAA ..................((.....))((((.(((....................(((....)))(((((......))))).............................))).)))).. ( -7.94 = -7.82 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:20 2006