| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,446,125 – 11,446,253 |
| Length | 128 |
| Max. P | 0.892964 |

| Location | 11,446,125 – 11,446,223 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11446125 98 - 20766785 UGGCUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCUGGAAACAGCUGUGGGCUGUCCC-CCCAC----------CCCU-UACCAUUCCCUUCUAAAGAGAAU (((.......))).(((((..............(((.((..((((....)))).(((((.......-)))))----------..))-.))).............))))). ( -23.43) >DroSec_CAF1 65006 98 - 1 UGGCUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCUGGAAACAGCUGUGGGCUGUCCC-CCCAC----------CCCU-UACCAUUCCCUUUUAAAGAAAAU ((((((((......................)))))))).((((((....)))).(((((.......-)))))----------....-.................)).... ( -22.15) >DroSim_CAF1 55010 99 - 1 UGCUUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCUGGAAACAGCUGUGGGCUGUCCCCCCCAC----------CCCU-UACCAUUCCCUUUUAAAGAAAAU .....................((((((.....(((......((((....)))).(((((........)))))----------....-.......))).......)))))) ( -20.70) >DroEre_CAF1 62048 108 - 1 UGGCUUCAUUCCAUUUUUCACAUUUUCUUUUCGGGUCAGUCGCUGGAAACAGCUGUGCGCUGUCCC-UCCACCUCCCUCUACCCCA-CCUCACUACCUUCUAAAGAGAAU (((.......)))........(((((((((..((((.(((.((((....)))).(((.(.......-.))))..............-....)))))))...))))))))) ( -20.20) >DroYak_CAF1 60701 102 - 1 UGGCUUUAUUCCGUUUUUCACAUUUUCUUUUCGGGUCAGUCGCUGAAAACAGCUGUGCG--------UCCACCUUCCUCUACCCCAACCCCCAUACCUUCUGAAGAGAAU ........................(((((((((((......((((....)))).(((..--------..)))..........................))))))))))). ( -16.40) >consensus UGGCUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCUGGAAACAGCUGUGGGCUGUCCC_CCCAC__________CCCU_UACCAUUCCCUUCUAAAGAGAAU ................................(((......((((....)))).(((((........)))))..........)))............((((....)))). (-15.72 = -15.40 + -0.32)

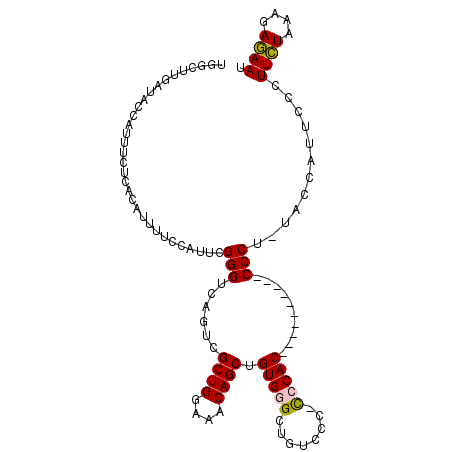

| Location | 11,446,152 – 11,446,253 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -20.98 |
| Energy contribution | -22.23 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
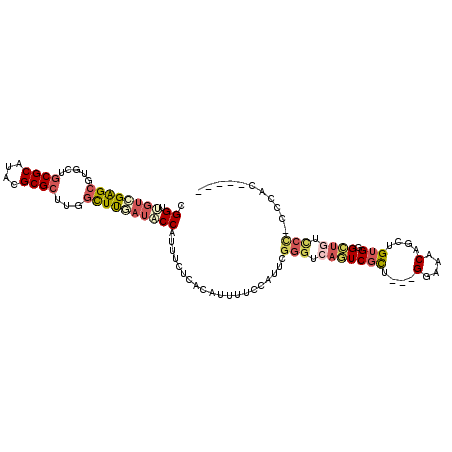
>2R_DroMel_CAF1 11446152 101 - 20766785 CGGUUGUCGAGCGUGCUGCGCAUACGCGCUUGGCUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCU---GGAAACAGCUGUGGGCUGUCCC-CCCAC----- .(((.(((((((.....((((....))))...))))))))))....................(((.((((((((---(....))))....))))).)))-.....----- ( -39.60) >DroSec_CAF1 65033 101 - 1 CGGUUGUCGAGCGUGCUGCGCAUACGCGCUUGGCUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCU---GGAAACAGCUGUGGGCUGUCCC-CCCAC----- .(((.(((((((.....((((....))))...))))))))))....................(((.((((((((---(....))))....))))).)))-.....----- ( -39.60) >DroSim_CAF1 55037 102 - 1 CGGUUGUCGAGCGUGCUGCGCAUACGCGCUUGCUUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCU---GGAAACAGCUGUGGGCUGUCCCCCCCAC----- .(((.((((((((....((((....)))).))).))))))))....................(((.((((((((---(....))))....))))).)))......----- ( -37.20) >DroEre_CAF1 62080 106 - 1 CGGUUGUCGAGCGUGCUGCGCAUACGCGCUUGGCUUCAUUCCAUUUUUCACAUUUUCUUUUCGGGUCAGUCGCU---GGAAACAGCUGUGCGCUGUCCC-UCCACCUCCC .(((.((((((((((.........))))))))))............................(((.((((.(((---(....)))).....)))).)))-...))).... ( -34.90) >DroYak_CAF1 60734 99 - 1 CGGUGGUCGAGCGUGCUGCGCAUACGCGCUUGGCUUUAUUCCGUUUUUCACAUUUUCUUUUCGGGUCAGUCGCU---GAAAACAGCUGUGCG--------UCCACCUUCC .(((((....(((.....)))..((((((..((((....((((..................))))..))))(((---(....)))).)))))--------)))))).... ( -31.37) >DroPer_CAF1 64918 84 - 1 AGGC-UCGGGGCGUGCUGCGCAUACGCGG-UGGUUCCAUUCCGUUU-------------UUCGGGUCACUCUUUUGGGAAAACAGCUGUGUGGU--UCU-UU-------- .((.-..(((((.(((((((....)))))-)))))))...))....-------------...(..((((.(..(((......)))..).)))).--.).-..-------- ( -25.80) >consensus CGGUUGUCGAGCGUGCUGCGCAUACGCGCUUGGCUUGAUACCAUUUCUCACAUUUUCCAUUCGGGUCAGUCGCU___GGAAACAGCUGUGGGCUGUCCC_CCCAC_____ .((.((((((((.....((((....))))...))))))))))....................(((.(((((((....(....)....))).)))).)))........... (-20.98 = -22.23 + 1.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:18 2006