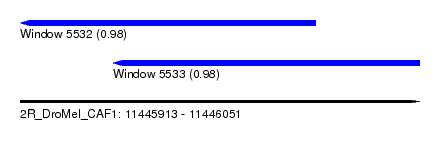
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,445,913 – 11,446,051 |
| Length | 138 |
| Max. P | 0.981034 |

| Location | 11,445,913 – 11,446,015 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -15.90 |
| Energy contribution | -17.76 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
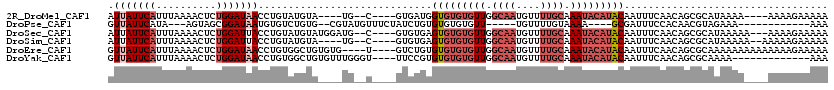
>2R_DroMel_CAF1 11445913 102 - 20766785 G--C----GUGAUGGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAA----AAAAGAAAAACAAAAGCAGGGCGGGCU---UU-----UUACCGUUCUCUU (--(----.(((..(((((((((.((((....)))).)))))))))....)))...))........----.............(((.(((((((...---..-----...)))))))))) ( -30.00) >DroSec_CAF1 64788 103 - 1 G--C----GUGUGAGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAAA---AAAAGAAAAACAAAAGCAGGGCGGGCU---UU-----UUACCGUUCUCUU (--(----((.((((((((((((.((((....)))).)))))))))....)))...)))).......---.............(((.(((((((...---..-----...)))))))))) ( -32.00) >DroSim_CAF1 54795 104 - 1 G--C----GUGUGAGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAAA--AAAAAGAAAAACAAAAGCAGGGCGGGCU---UU-----UUACCGUUCUCUU (--(----((.((((((((((((.((((....)))).)))))))))....)))...)))).......--..............(((.(((((((...---..-----...)))))))))) ( -32.00) >DroEre_CAF1 61830 105 - 1 ---U----GUCUGUGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAAAAAAAAAAAAAAGAAAAACAAAAGCAGGGCGGGCU---UU-----UUACCGUUCUCUU ---(----(((((((((((((((.((((....)))).))))))))).......)))).)))......................(((.(((((((...---..-----...)))))))))) ( -27.41) >DroYak_CAF1 60487 97 - 1 GGGU----UUCCGUGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAAAA-------------AAACAAAAGCAGGGCGGGCU---UUUUUUUUUACCGUUCU--- ..((----(....((((((((((.((((....)))).)))))))))).....))).((......-------------........))(((((((...---..........)))))))--- ( -25.06) >DroPer_CAF1 64668 93 - 1 AUGUUUCUAUCUGUGUGUGUGUU-----UGUUUUGUAAAA----GCGAUUUCCACAACGUAGAAA------------AAACAGAAGCAGGGCAGGGCUCCCU-----UUCACGUUCC-GU .(((((((.(((((((.((((.(-----(((((.....))----))))....)))))))))))..------------....))))))).((.(((....)))-----.)).......-.. ( -18.70) >consensus G__C____GUCUGUGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAA____AAAAGAAAAACAAAAGCAGGGCGGGCU___UU_____UUACCGUUCUCUU ..............(((((((((.((((....)))).))))))))).........................................(((((((................)))))))... (-15.90 = -17.76 + 1.86)

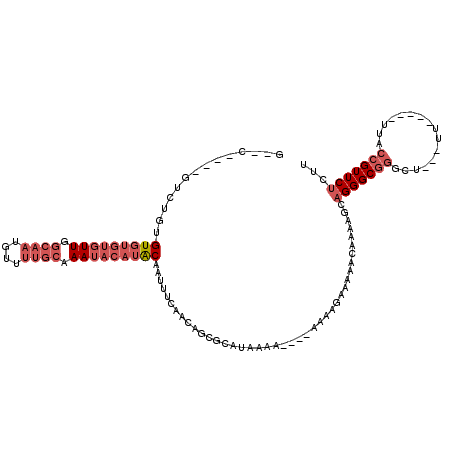
| Location | 11,445,945 – 11,446,051 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.05 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -11.22 |
| Energy contribution | -13.08 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11445945 106 - 20766785 AUUAUUCAUUUAAAACUCUGGAUAACCUGUAUGUA----UG--C----GUGAUGGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAA----AAAAGAAAAA .(((((((..........))))))).((.....((----((--(----((((..(((((((((.((((....)))).)))))))))....))....)))))))...----...))..... ( -25.70) >DroPse_CAF1 61924 94 - 1 GUUAUUCAUA---AGUAGCGGAUAAUGUGUCUGUG--CGUAUGUUUCUAUCUGUGUGUGUGUU-----UGUUUUGUAAAA----GCGAUUUCCACAACGUAGAAA------------AAA ......((((---.(((.(((((.....)))))))--).))))(((((((...((((.(...(-----(((((.....))----))))...)))))..)))))))------------... ( -21.10) >DroSec_CAF1 64820 111 - 1 AUUAUUCAUUUAAAACUCUGGAUUACCUGUAUGUAUGGAUG--C----GUGUGAGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAAA---AAAAGAAAAA ....(((..................((.........))(((--(----((.((((((((((((.((((....)))).)))))))))....)))...)))))).....---....)))... ( -25.30) >DroSim_CAF1 54827 108 - 1 AUUAUUCAUUUAAAACUCUGGAUUACCUGUAUGUA----UG--C----GUGUGAGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAAA--AAAAAGAAAAA ....(((.......((...((....)).))...((----((--(----((.((((((((((((.((((....)))).)))))))))....)))...)))))))....--.....)))... ( -26.20) >DroEre_CAF1 61862 112 - 1 GUUAUUCAUUUAAAACUCUGGAUAACCUGUGGCUGUGUG----U----GUCUGUGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAAAAAAAAAAAAAAGAAAAA ((((((((..........)))))))).((((.((((.((----.----.....((((((((((.((((....)))).))))))))))....))))))))))................... ( -29.30) >DroYak_CAF1 60521 103 - 1 GUUAUUCAUUUAAAACUCUGGAUAACCUGUGGCUGUGUUUGGGU----UUCCGUGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAAAA-------------AAA ((((((((..........)))))))).((((.((((....((..----..)).((((((((((.((((....)))).))))))))))......))))))))...-------------... ( -29.50) >consensus AUUAUUCAUUUAAAACUCUGGAUAACCUGUAUGUA____UG__C____GUCUGUGUGUGUGUUGGCAAUGUUUUGCAAAUACAUACAAUUUCAACAGCGCAUAAAA____AAAAGAAAAA .(((((((..........))))))).............................(((((((((.((((....)))).))))))))).................................. (-11.22 = -13.08 + 1.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:16 2006