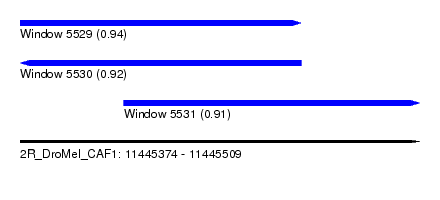
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,445,374 – 11,445,509 |
| Length | 135 |
| Max. P | 0.936644 |

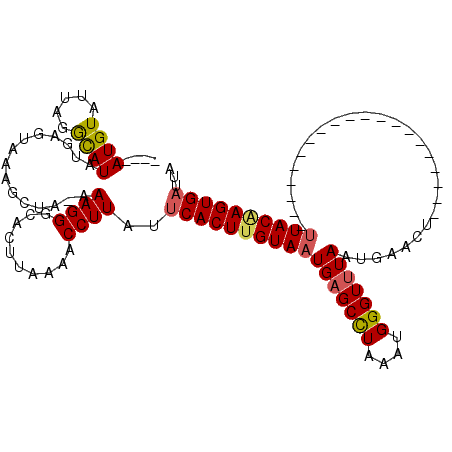
| Location | 11,445,374 – 11,445,469 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
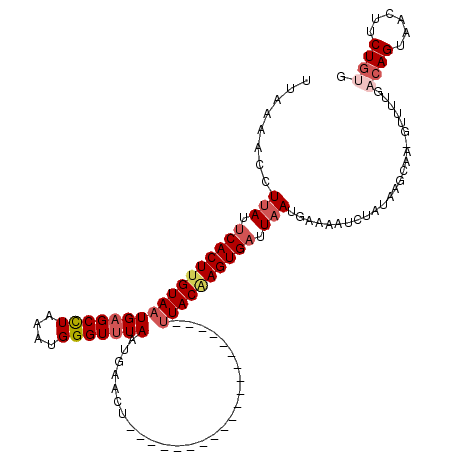
>2R_DroMel_CAF1 11445374 95 + 20766785 ----AUGUAUUAGGCAUAUGAGUUAAGCUA-AAAGGGCACUUAAAACCUUAAUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU--------------------UUACAAGUGAUUA ----.......(((....(((((...(((.-....))))))))...)))((((((((((((((((((((....))))))).......--------------------))))))))))))) ( -24.51) >DroSec_CAF1 64254 95 + 1 ----AUGUAUUAGGCAUAUGAGUUAAGCUA-AAAGGGCACUUAAAACCUUAUUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU--------------------UUACGAGUGAUUA ----.......(((....(((((...(((.-....))))))))...)))...(((((((((((((((((....))))))).......--------------------))))))))))... ( -21.51) >DroSim_CAF1 54263 95 + 1 ----AUGUAUUAGACAUAUGAGUAAAGCUA-AAAGGUCACUUAAAACCUUAUUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU--------------------UUACAAGUGAUUA ----........(((.....((.....)).-....)))..............(((((((((((((((((....))))))).......--------------------))))))))))... ( -20.81) >DroEre_CAF1 61268 119 + 1 AGCAAUGUAUCAGGUAUAUGAAUAAGGCUUAAAAGGCUAUUUAAAACCUUAUUCACUUGUAAUGCGCUUAAAUGGGUUUAAUGAACUCCGCGAAUGG-GGCAGCGAAUUACAAGUGAUUA ...........((((...(((((..((((.....)))))))))..))))...(((((((((((.((((.....((((((...)))).))((......-.)))))).)))))))))))... ( -30.40) >DroYak_CAF1 59935 120 + 1 AGAAAUGUAUAAGGCAUAUGAAUAAAACGAAAAAGGGCACUUAAAACCUUAUUCACUCGUAAUGAGCUUUAAUGGGUUUAAUGAACUCCACAAAUGGGGGCAGCGAAAUACAAGUGUGUU ............(((((((.........(((.((((..........)))).)))..((((..(((((((....))))))).....(((((....)))))...)))).......))))))) ( -23.10) >consensus ____AUGUAUUAGGCAUAUGAGUAAAGCUA_AAAGGGCACUUAAAACCUUAUUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU____________________UUACAAGUGAUUA ....((((.....))))...............((((..........))))..(((((((((((((((((....)))))))...........................))))))))))... (-14.52 = -14.64 + 0.12)

| Location | 11,445,374 – 11,445,469 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -8.28 |
| Energy contribution | -9.24 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11445374 95 - 20766785 UAAUCACUUGUAA--------------------AGUUCAUUAAACCCAUUUAGGCUCAUUACAAGUGAUUAAGGUUUUAAGUGCCCUUU-UAGCUUAACUCAUAUGCCUAAUACAU---- (((((((((((((--------------------.(....(((((....)))))...).)))))))))))))((((.((((((.......-..)))))).......)))).......---- ( -19.80) >DroSec_CAF1 64254 95 - 1 UAAUCACUCGUAA--------------------AGUUCAUUAAACCCAUUUAGGCUCAUUACAAGUGAAUAAGGUUUUAAGUGCCCUUU-UAGCUUAACUCAUAUGCCUAAUACAU---- .........(((.--------------------.(((.....)))....((((((((((.....))))...(((((..(((....))).-.))))).........)))))))))..---- ( -13.00) >DroSim_CAF1 54263 95 - 1 UAAUCACUUGUAA--------------------AGUUCAUUAAACCCAUUUAGGCUCAUUACAAGUGAAUAAGGUUUUAAGUGACCUUU-UAGCUUUACUCAUAUGUCUAAUACAU---- ...((((((((((--------------------.(....(((((....)))))...).)))))))))).(((((((..(((....))).-.)))))))..................---- ( -17.90) >DroEre_CAF1 61268 119 - 1 UAAUCACUUGUAAUUCGCUGCC-CCAUUCGCGGAGUUCAUUAAACCCAUUUAAGCGCAUUACAAGUGAAUAAGGUUUUAAAUAGCCUUUUAAGCCUUAUUCAUAUACCUGAUACAUUGCU ..((((((((((((.((((((.-......))((.(((.....))))).....)))).)))))))))(((((((((((.(((......))))))))))))))........)))........ ( -32.40) >DroYak_CAF1 59935 120 - 1 AACACACUUGUAUUUCGCUGCCCCCAUUUGUGGAGUUCAUUAAACCCAUUAAAGCUCAUUACGAGUGAAUAAGGUUUUAAGUGCCCUUUUUCGUUUUAUUCAUAUGCCUUAUACAUUUCU ....((((((((....(((..........((((.(((.....)))))))...)))....)))))))).(((((((..............................)))))))........ ( -20.83) >consensus UAAUCACUUGUAA____________________AGUUCAUUAAACCCAUUUAGGCUCAUUACAAGUGAAUAAGGUUUUAAGUGCCCUUU_UAGCUUUACUCAUAUGCCUAAUACAU____ ...((((((((((.............................................)))))))))).(((((((..(((....)))...)))))))...................... ( -8.28 = -9.24 + 0.96)

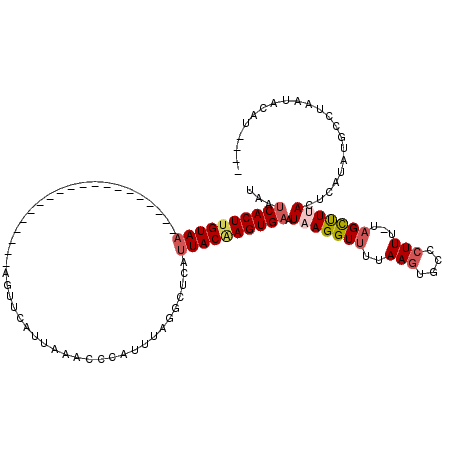
| Location | 11,445,409 – 11,445,509 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -12.98 |
| Energy contribution | -14.22 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11445409 100 + 20766785 UUAAAACCUUAAUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU--------------------UUACAAGUGAUUAAUCAAAAUCUAUAAGCAAUGUUUUGACAGUAACUUCUGUG ........(((((((((((((((((((((....))))))).......--------------------)))))))))))))).((((((...........))))))((((......)))). ( -24.61) >DroSec_CAF1 64289 100 + 1 UUAAAACCUUAUUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU--------------------UUACGAGUGAUUAAUGAAAAUCUAUAAGCAAAGUUUUGUCAGUAACUUCUGUG ......(.(((.(((((((((((((((((....))))))).......--------------------)))))))))).))).)...........((((((((........))))).))). ( -21.01) >DroSim_CAF1 54298 100 + 1 UUAAAACCUUAUUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU--------------------UUACAAGUGAUUAAUGAAAAGCUACAAGCAAAGUUUUGACAGUAACUUCUGUG .((((((.(((.(((((((((((((((((....))))))).......--------------------)))))))))).)))......((.....))...))))))((((......)))). ( -24.81) >DroEre_CAF1 61308 115 + 1 UUAAAACCUUAUUCACUUGUAAUGCGCUUAAAUGGGUUUAAUGAACUCCGCGAAUGG-GGCAGCGAAUUACAAGUGAUUAAUGCAAAUCUGUAAG----GGGUUGACAGUAACUUCUGUG ....((((((..(((((((((((.((((.....((((((...)))).))((......-.)))))).)))))))))))....((((....)))).)----))))).((((......)))). ( -33.10) >DroYak_CAF1 59975 116 + 1 UUAAAACCUUAUUCACUCGUAAUGAGCUUUAAUGGGUUUAAUGAACUCCACAAAUGGGGGCAGCGAAAUACAAGUGUGUUAUGCAAAUCUGUAAG----GUUUUGACAGUAACUGCUUCG ((((((((((((....((((..(((((((....))))))))))).(((((....)))))...(((.(((((....))))).)))......)))))----))))))).............. ( -28.00) >consensus UUAAAACCUUAUUCACUUGUAAUGAGCCUAAAUGGGUUUAAUGAACU____________________UUACAAGUGAUUAAUGAAAAUCUAUAAGCAA_GUUUUGACAGUAACUUCUGUG ........(((.(((((((((((((((((....)))))))...........................)))))))))).)))........................((((......)))). (-12.98 = -14.22 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:14 2006