
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,444,096 – 11,444,244 |
| Length | 148 |
| Max. P | 0.665978 |

| Location | 11,444,096 – 11,444,214 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11444096 118 + 20766785 GCAUGUGGAAAUGUUGUGUUCAAUCAAAAGCAGCACAUAGUUGCCAUAGAAACCGGCAAACCUAACUUGCAUAAGUUUAUGCAGUUUAUUAACUGUAUAACGGUUAUACAUCUAAGCA ((.(((((.(((..(((((((........).))))))..))).)))))..(((((((((.......))))......((((((((((....)))))))))))))))..........)). ( -32.30) >DroSec_CAF1 63000 118 + 1 GCACGUGGAAAUGUUGUGUUUAAUCAAAAGCAGCACAUAGUUGCCAUAGAAACCGGCAAACUUAACUUGCAUAAGUUUAUGCAGUUUGUAAUCUGUAAAGCGGUUAUACAUCUAAGCA ((((((....)))).((((.(((((....(((((.....))))).(((((.....(((((((......(((((....))))))))))))..))))).....))))).))))....)). ( -28.40) >DroSim_CAF1 53006 118 + 1 GCAUGUGGAACUGUUGUGUUUAUUCAAAAGCAGCACAUAGUUGCCAUAGAAACCGGCAAACUUAACUUGCAUAAGUUCAUGCAGUUUGUUAACUGUAUAACGGUUAUACAUCUAAGCA ((.(((((((((((.(((((...........))))))))))).)))))..(((((((((.......))))........((((((((....))))))))..)))))..........)). ( -33.00) >DroEre_CAF1 59934 118 + 1 GCAUGUGGCAAUGUUGUGUUUAAUCAAAAGCAGCACAUAGUAGUCAUGGAAACCGGCGAGCUCAACUUGCAUAAUUUCGUGCAGUUUGUUAACUGUUUACCGGUAAUGCAUCUAAGGA (((((((((.((..((((((...........))))))..)).)))))....(((((..(((.(((.((((((......)))))).)))......)))..))))).))))......... ( -31.20) >DroYak_CAF1 58645 118 + 1 GCAUGUGGCAAUGUUGUGUUUAAUCAAAAGCGGCACACAGUAGUCAUGGCAACCGGCGAACUCAACUUGCAUAAGUUCGUGCAGUUUGUUUGCUGUUCAGCGGUUAUGCAUCUAAGCA ((.((((((....(((........))).....)).)))).(((.((((((..(((((((((..((((.((((......)))))))).))))))))....)..))))))...))).)). ( -31.50) >consensus GCAUGUGGAAAUGUUGUGUUUAAUCAAAAGCAGCACAUAGUUGCCAUAGAAACCGGCAAACUUAACUUGCAUAAGUUCAUGCAGUUUGUUAACUGUAUAACGGUUAUACAUCUAAGCA ((.(((((......((((((...........))))))......)))))..(((((((((.......))))........((((((((....))))))))..)))))..........)). (-22.44 = -22.84 + 0.40)

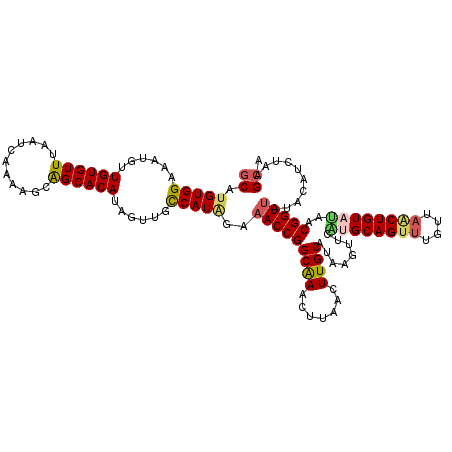
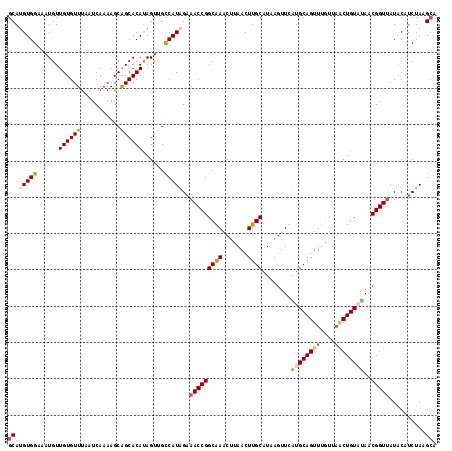
| Location | 11,444,134 – 11,444,244 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11444134 110 + 20766785 AGUUGCCAUAGAAACCGGCAAACCUAACUUGCAUAAGUUUAUGCAGUUUAUUAACUGUAUAACGGUUAUACAUCUAAGCAAAC---------UUUUUU-AAUUUUAAGGAUUCUAUAAGA ..((((..(((((((((((((.......))))......((((((((((....))))))))))))))).....)))).))))..---------......-..................... ( -23.00) >DroSec_CAF1 63038 110 + 1 AGUUGCCAUAGAAACCGGCAAACUUAACUUGCAUAAGUUUAUGCAGUUUGUAAUCUGUAAAGCGGUUAUACAUCUAAGCAAAC---------UUUUGU-GAUUUUAAGGAUUCUAUGAAA ......(((((((.((.(((((((......(((((....))))))))))))((((...((((..(((.........)))...)---------)))...-))))....)).)))))))... ( -22.60) >DroSim_CAF1 53044 98 + 1 AGUUGCCAUAGAAACCGGCAAACUUAACUUGCAUAAGUUCAUGCAGUUUGUUAACUGUAUAACGGUUAUACAUCUAAGCAAAC---------UUUUGU-GAUUUUAAG------------ ..(((((.........))))).(((((.(..((.(((((..(((....(((((((((.....)))))).))).....))))))---------)).)).-.)..)))))------------ ( -22.30) >DroEre_CAF1 59972 117 + 1 AGUAGUCAUGGAAACCGGCGAGCUCAACUUGCAUAAUUUCGUGCAGUUUGUUAACUGUUUACCGGUAAUGCAUCUAAGGAAUCUAACUAAGGAAUGCAAGUAGAUAUGUA---UAUACAA .(((..((((...(((((..(((.(((.((((((......)))))).)))......)))..)))))..(((((((.((........)).))..)))))......))))..---..))).. ( -27.20) >DroYak_CAF1 58683 107 + 1 AGUAGUCAUGGCAACCGGCGAACUCAACUUGCAUAAGUUCGUGCAGUUUGUUUGCUGUUCAGCGGUUAUGCAUCUAAGCAAAC---------AUUGCU-GUUUUUAGAAA---UAUGAAA .....(((((.(...((((((((..((((.((((......)))))))).))))))))..(((((((..(((......)))...---------))))))-)......)...---))))).. ( -29.10) >consensus AGUUGCCAUAGAAACCGGCAAACUUAACUUGCAUAAGUUCAUGCAGUUUGUUAACUGUAUAACGGUUAUACAUCUAAGCAAAC_________UUUUCU_GAUUUUAAGGA___UAUAAAA ..((((..(((((((((((((.......))))........((((((((....))))))))..))))).....)))).))))....................................... (-14.94 = -15.82 + 0.88)

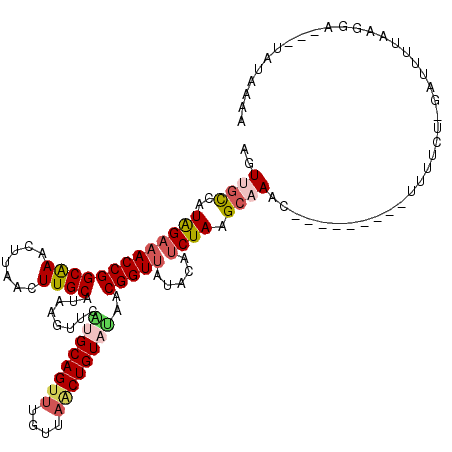
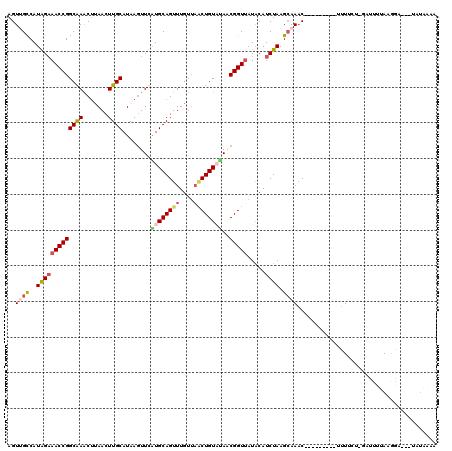
| Location | 11,444,134 – 11,444,244 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -12.06 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11444134 110 - 20766785 UCUUAUAGAAUCCUUAAAAUU-AAAAAA---------GUUUGCUUAGAUGUAUAACCGUUAUACAGUUAAUAAACUGCAUAAACUUAUGCAAGUUAGGUUUGCCGGUUUCUAUGGCAACU ..((((((((.((........-.....(---------(((((.((((.((((((.....)))))).))))))))))(((((....)))))......((....)))).))))))))..... ( -23.30) >DroSec_CAF1 63038 110 - 1 UUUCAUAGAAUCCUUAAAAUC-ACAAAA---------GUUUGCUUAGAUGUAUAACCGCUUUACAGAUUACAAACUGCAUAAACUUAUGCAAGUUAAGUUUGCCGGUUUCUAUGGCAACU ..((((((((.((........-.....(---------(((((...(..((((.........))))..)..))))))(((..((((((.......))))))))).)).))))))))..... ( -22.30) >DroSim_CAF1 53044 98 - 1 ------------CUUAAAAUC-ACAAAA---------GUUUGCUUAGAUGUAUAACCGUUAUACAGUUAACAAACUGCAUGAACUUAUGCAAGUUAAGUUUGCCGGUUUCUAUGGCAACU ------------(((((....-.....(---------(((((.((((.((((((.....)))))).))))))))))(((((....)))))...))))).((((((.......)))))).. ( -22.90) >DroEre_CAF1 59972 117 - 1 UUGUAUA---UACAUAUCUACUUGCAUUCCUUAGUUAGAUUCCUUAGAUGCAUUACCGGUAAACAGUUAACAAACUGCACGAAAUUAUGCAAGUUGAGCUCGCCGGUUUCCAUGACUACU .(((((.---.....(((((((..........)).))))).......)))))..((((((....((((....(((((((........))).)))).)))).))))))............. ( -20.82) >DroYak_CAF1 58683 107 - 1 UUUCAUA---UUUCUAAAAAC-AGCAAU---------GUUUGCUUAGAUGCAUAACCGCUGAACAGCAAACAAACUGCACGAACUUAUGCAAGUUGAGUUCGCCGGUUGCCAUGACUACU ..((((.---...........-.....(---------(((((((.....((......)).....))))))))(((((..((((((((.......)))))))).)))))...))))..... ( -25.40) >consensus UUUCAUA___UCCUUAAAAUC_ACAAAA_________GUUUGCUUAGAUGUAUAACCGCUAUACAGUUAACAAACUGCAUGAACUUAUGCAAGUUAAGUUUGCCGGUUUCUAUGGCAACU ...............................................(((...(((((.....(((((....)))))..((((((((.......)))))))).)))))..)))....... (-12.06 = -11.90 + -0.16)

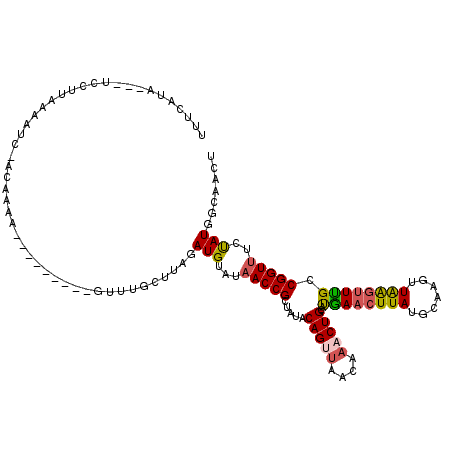
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:12 2006