| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,443,521 – 11,443,637 |
| Length | 116 |
| Max. P | 0.773251 |

| Location | 11,443,521 – 11,443,612 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.99 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
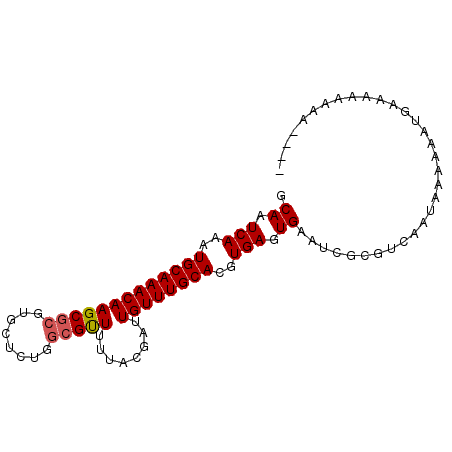
>2R_DroMel_CAF1 11443521 91 + 20766785 AUCACUUUUCAUUCUACGUUAAAAUGCAAUCAAAUGCAAACAAGCGCGUGCUCUGG--CGUUUUUACGAU-UGUUUGCACGUGAGUGAAUC----GCG .......(((((((.((((......((((((((((((.....(((....)))...)--)))))....)))-)))....)))))))))))..----... ( -22.00) >DroVir_CAF1 80221 92 + 1 UUUUCUUCGCAUGAU--AGCCAAGUGCAAUCAAAUGCAAACAAGCGCGUGCUUACAGC----UUUAUGAUAUGUUUGCACGUGAGUGGACCCUAGGCG ...............--.(((.((((((......))))..((..(((((((..(((.(----.....)...)))..)))))))..))....)).))). ( -26.50) >DroPse_CAF1 59824 90 + 1 AUCAGCUGGCGUUAUAUGCAAAAAUGCAAUCAAAUGCAAACAAGCGCGUGCUCUCA---GCUUUUACGAU-UGUUUGCACGUGAGUGAAUC----GCG .(((.((.((((....((((....)))).......((((((((...(((((.....---))....))).)-))))))))))).)))))...----... ( -24.30) >DroEre_CAF1 59351 91 + 1 AUCACUUUGCAUUAAAUGGAAAAAUGCAAUCAAAUGCAAACAAGCGCGUGCUCUGG--CGUUUUUACGAU-UGUUUGCACGUGAGUGAAUC----GCG .((((.(((((((.........)))))))(((..(((((((((((((........)--)))........)-))))))))..)))))))...----... ( -26.00) >DroAna_CAF1 42528 89 + 1 AUCAGCUCGCAUUAAGUGCAAAAAUGCAAUCAAAUGCAAACAAGCGCGUGCUCUGG--CGUU--UAUGGA-UGUUUGCACGUGAGUGCGCC----GCU ...(((.((((((..(((((((..((((......))))...((((((........)--))))--).....-..)))))))...))))))..----))) ( -29.10) >DroPer_CAF1 62617 90 + 1 AUCAGCUGGCGUUAUAUGCAAAAAUGCAAUCAAAUGCAAACAAGCGCGUGCUCUCA---GCUUUUACGAU-UGUUUGCACGUGAGUGAAUC----GCG .(((.((.((((....((((....)))).......((((((((...(((((.....---))....))).)-))))))))))).)))))...----... ( -24.30) >consensus AUCACCUCGCAUUAUAUGCAAAAAUGCAAUCAAAUGCAAACAAGCGCGUGCUCUCA___GUUUUUACGAU_UGUUUGCACGUGAGUGAAUC____GCG ........((..............((((......))))..((..(((((((.........................)))))))..))........)). (-16.83 = -16.99 + 0.17)


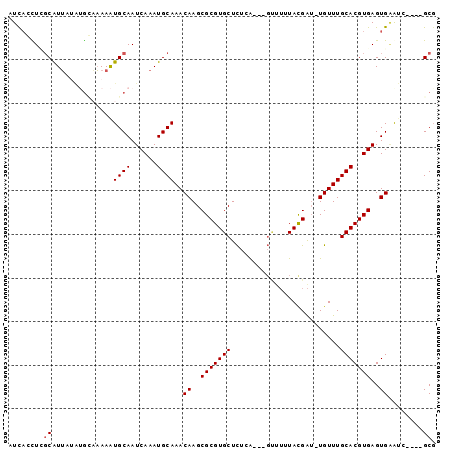
| Location | 11,443,521 – 11,443,612 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11443521 91 - 20766785 CGC----GAUUCACUCACGUGCAAACA-AUCGUAAAAACG--CCAGAGCACGCGCUUGUUUGCAUUUGAUUGCAUUUUAACGUAGAAUGAAAAGUGAU ...----...(((((((.(((((((((-(.(((......(--(....))..))).)))))))))).)))...(((((((...)))))))....)))). ( -22.80) >DroVir_CAF1 80221 92 - 1 CGCCUAGGGUCCACUCACGUGCAAACAUAUCAUAAA----GCUGUAAGCACGCGCUUGUUUGCAUUUGAUUGCACUUGGCU--AUCAUGCGAAGAAAA .(((...(((.((.(((.(((((((((........(----(((((....))).)))))))))))).))).)).))).))).--............... ( -23.40) >DroPse_CAF1 59824 90 - 1 CGC----GAUUCACUCACGUGCAAACA-AUCGUAAAAGC---UGAGAGCACGCGCUUGUUUGCAUUUGAUUGCAUUUUUGCAUAUAACGCCAGCUGAU .((----(......(((.(((((((((-(.(((....((---.....))..))).)))))))))).))).((((....)))).....)))........ ( -25.90) >DroEre_CAF1 59351 91 - 1 CGC----GAUUCACUCACGUGCAAACA-AUCGUAAAAACG--CCAGAGCACGCGCUUGUUUGCAUUUGAUUGCAUUUUUCCAUUUAAUGCAAAGUGAU ...----...(((((((.(((((((((-(.(((......(--(....))..))).)))))))))).)))(((((((.........))))))).)))). ( -25.90) >DroAna_CAF1 42528 89 - 1 AGC----GGCGCACUCACGUGCAAACA-UCCAUA--AACG--CCAGAGCACGCGCUUGUUUGCAUUUGAUUGCAUUUUUGCACUUAAUGCGAGCUGAU ..(----((((((.(((.(((((((((-......--..((--(........)))..))))))))).))).)))....(((((.....))))))))).. ( -28.00) >DroPer_CAF1 62617 90 - 1 CGC----GAUUCACUCACGUGCAAACA-AUCGUAAAAGC---UGAGAGCACGCGCUUGUUUGCAUUUGAUUGCAUUUUUGCAUAUAACGCCAGCUGAU .((----(......(((.(((((((((-(.(((....((---.....))..))).)))))))))).))).((((....)))).....)))........ ( -25.90) >consensus CGC____GAUUCACUCACGUGCAAACA_AUCGUAAAAAC___CCAGAGCACGCGCUUGUUUGCAUUUGAUUGCAUUUUUGCAUAUAAUGCAAACUGAU .......(((.((.(((.((((((((.....((....))......((((....)))))))))))).))).)).)))...................... (-14.36 = -14.37 + 0.00)

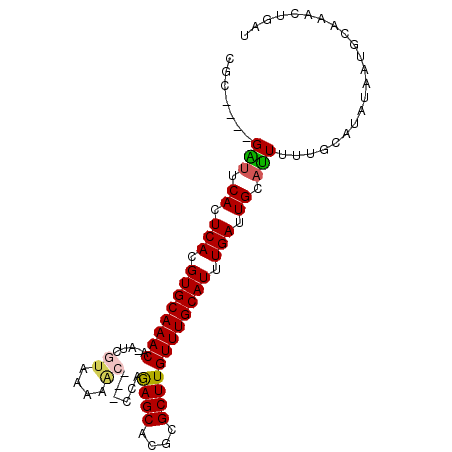
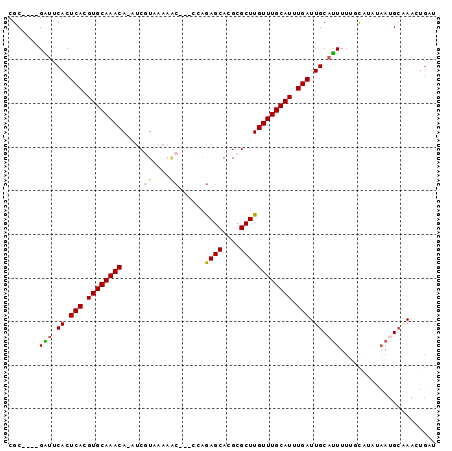
| Location | 11,443,546 – 11,443,637 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11443546 91 + 20766785 GCAAUCAAAUGCAAACAAGCGCGUGCUCUGGCGUUUUUACGAUUGUUUGCACGUGAGUGAAUCGCGUCUAUAAAAAAUGAAAAAAAAAAAA ....(((...((((((((((((........))))........))))))))((((((.....))))))..........)))........... ( -20.20) >DroSec_CAF1 62475 91 + 1 GCAAUCAAAUGCAAACAAGCGCGUGCUCUGGCGUUUUUACGAUUGUUUGCACGUGAGUGAAUCGCGUCUAUAAAAAAUGAAAAAAAAAAAA ....(((...((((((((((((........))))........))))))))((((((.....))))))..........)))........... ( -20.20) >DroEre_CAF1 59376 87 + 1 GCAAUCAAAUGCAAACAAGCGCGUGCUCUGGCGUUUUUACGAUUGUUUGCACGUGAGUGAAUCGCGACAAUAAAAAAUAUGAAAAAA---- ....(((..(((((((((((((........))))........)))))))))(((((.....))))).............))).....---- ( -20.40) >DroYak_CAF1 58088 86 + 1 GCAAUCAAAUGCAAACAAGCGCGUGCUCUGGCGUUUUUACGAUUGUUUGCACGUGAGUGAAUCGCGUCAAGAAAAAAUAUGAAAAA----- ....(((...((((((((((((........))))........))))))))((((((.....))))))............)))....----- ( -20.20) >DroAna_CAF1 42553 88 + 1 GCAAUCAAAUGCAAACAAGCGCGUGCUCUGGCGUU--UAUGGAUGUUUGCACGUGAGUGCGCCGCUCCGCAAACAAGUGCCAGCAAAAAC- (((.(((..(((((((((((((........)))))--......))))))))..))).)))...(((..(((......))).)))......- ( -26.50) >DroPer_CAF1 62642 75 + 1 GCAAUCAAAUGCAAACAAGCGCGUGCUCUCA-GCUUUUACGAUUGUUUGCACGUGAGUGAAUCGCGUCCGCCAACA--------------- ((........((((((((...(((((.....-))....))).))))))))((((((.....))))))..)).....--------------- ( -20.90) >consensus GCAAUCAAAUGCAAACAAGCGCGUGCUCUGGCGUUUUUACGAUUGUUUGCACGUGAGUGAAUCGCGUCAAUAAAAAAUGAAAAAAAA____ .((.(((..(((((((((((((........)))))........))))))))..))).))................................ (-16.66 = -16.85 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:09 2006