
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,442,986 – 11,443,200 |
| Length | 214 |
| Max. P | 0.744657 |

| Location | 11,442,986 – 11,443,085 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.09 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -9.60 |
| Energy contribution | -10.12 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11442986 99 + 20766785 UAAAUGGUGACUAAUUUGAGAAUGACUAAUCUGCAUUUAAUUAACCGAUUGAUUGAACGGAUUUCUCAAUUUAGUGUUUU-----------UUGUUUGUUAAAAAAUA-AA--------- .........(((((.(((((((......(((((..(((((((((....))))))))))))))))))))).)))))(((((-----------(((.....)))))))).-..--------- ( -20.70) >DroSec_CAF1 61933 95 + 1 UAAAUGGUGACUAAUUUGAGAAUGACUAAUCUGCAUUAAAUUAACCGAUUGAUUGAACGGAUUUCUCAAUUUAGU----------------UAAAUAACUAACAAAUAUAG--------- .......(((((((.(((((((......(((((.....((((((....))))))...)))))))))))).)))))----------------))..................--------- ( -17.30) >DroSim_CAF1 51931 95 + 1 UAAAUGGUGACUAAUUUGAGAAUGACUAAUCUGCAUUUAAUUAACCGAUUGAUUGAACGGAUUUCUCAAUUUAGU----------------UAAAUAACUAACAAAAGUAA--------- .......(((((((.(((((((......(((((..(((((((((....))))))))))))))))))))).)))))----------------))..................--------- ( -20.10) >DroEre_CAF1 58819 111 + 1 UAAAUGGUGACUAAUUUGAGAAUGACUAAUGUUCAUUUGGUUA-CCGAUCGAUUGAACGGAUUUAUCAAUUUAGUGUUUUUUUUUAU-UUUUAA----CCAACAAAAGCA---AGUCAAU ....((((....(((..(((((..(((((((((((.((((((.-..)))))).))))))((....))...)))))...)))))..))-)....)----))).........---....... ( -18.40) >DroYak_CAF1 57519 119 + 1 UUAAUGGUGACUAAUUUACGAAUGACUAAUGUUCAUUUGGUUAACCGUUCGAUUAAACGGAUUUCUCCGUUUAGUGUUUUU-CUUAUUUGUUAACUAACUAACAAAAGUCAACAGUCAAU ....((.(((((......(((((((((((.......)))))....))))))((((((((((....))))))))))......-....(((((((......)))))))))))).))...... ( -28.00) >consensus UAAAUGGUGACUAAUUUGAGAAUGACUAAUCUGCAUUUAAUUAACCGAUUGAUUGAACGGAUUUCUCAAUUUAGUGUUUU___________UAAAUAACUAACAAAAGUAA_________ .........(((((.(((((((...((......((.((((((....)))))).))...))..))))))).)))))............................................. ( -9.60 = -10.12 + 0.52)

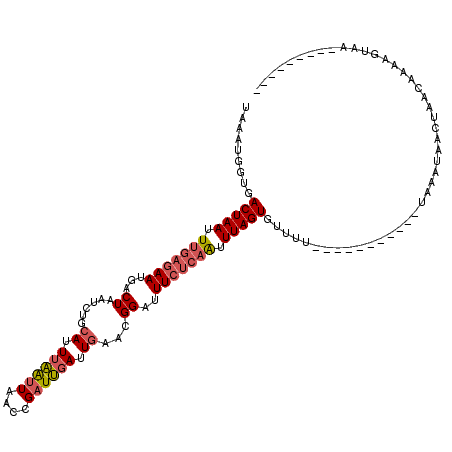
| Location | 11,443,085 – 11,443,200 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11443085 115 + 20766785 ----ACAGCUACUAAAACCGCCAAACUAGCUUUUACUAAUUCAAACGAGUUAUUUCGUU-GAAAGUAUACAACUUAAUAACCGAAUCGGAAAACGAUAAUUAGCGAACAAGUAACUCAAU ----..(((((...............)))))...............(((((((((((((-((((((.....)))).........((((.....))))..))))))))...)))))))... ( -21.06) >DroSec_CAF1 62028 115 + 1 ----GCACCUACUAAAACGGUCAAACUAGCUAGUACUAAUUCAAACGAGUUAUUUCGUU-GAAAGUAUACAAGUUAAUAACCGAAUCGGUAAACGAUAAUUAGCGAACAAGAAUCUCAAU ----((...........((((.....(((((.(((((..(((((.((((....))))))-))))))))...)))))...)))).((((.....)))).....))................ ( -21.60) >DroSim_CAF1 52026 115 + 1 ----GCAGCUACUAAAACGACCAAACUAGCUAGUACUAAUUCAAACGAGUUAUUUUGUU-GAAAGUAUACAAGUUAAUAACCGAAUCGGAAAACGAUAAUUAGCGAACAAGAAUCUCAAU ----...((((...............(((((.(((((..(((((.((((....))))))-))))))))...)))))........((((.....))))...))))................ ( -17.00) >DroEre_CAF1 58930 119 + 1 UGAUGUGGCUUCUAAAACGGGCA-ACUAGCUAGUACUAAUUCAAACGAGUUCUUUCGUUAGAAAGUAUACAACUUAAUAACCGAAUCGAAAAACGAUAAUUAGCGAACAAGAAUCUCAAU ...((.((.((((.....((...-.)).(((((((((..(((.((((((....)))))).))))))))................((((.....))))...)))).....)))))).)).. ( -21.70) >DroYak_CAF1 57638 119 + 1 UGAUGUGGCUACUAAAACGGGCAAACUAGCUGGUACUAAUUCAAACGAGUUAUUUCGUUAGAAAGUAUACAACUUAAUAACCGAAUCGAAAAACGAUAAUUAGCGAACAAGAAU-UCCAU ...(((.((((......(((.......((.(((((((..(((.((((((....)))))).)))))))).)).))......))).((((.....))))...))))..))).....-..... ( -22.92) >consensus ____GCAGCUACUAAAACGGCCAAACUAGCUAGUACUAAUUCAAACGAGUUAUUUCGUU_GAAAGUAUACAACUUAAUAACCGAAUCGGAAAACGAUAAUUAGCGAACAAGAAUCUCAAU ............................(((((((((..(((.((((((....)))))).))))))))................((((.....))))...))))................ (-15.66 = -15.78 + 0.12)

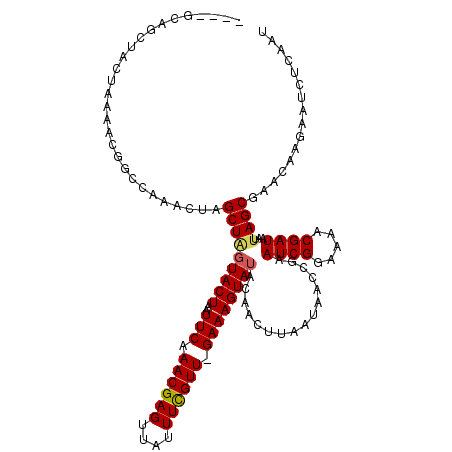

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:06 2006