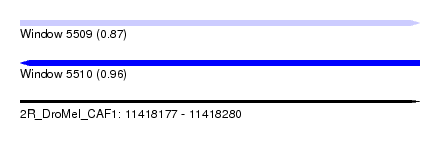
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,418,177 – 11,418,280 |
| Length | 103 |
| Max. P | 0.957600 |

| Location | 11,418,177 – 11,418,280 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -18.15 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11418177 103 + 20766785 AA---UAAAGGGGGUAACAAGCGGUGGGUUACGGGUAAUGGGUAGCGGCAAGCGGAGGACCUGGAGUGGACCGCACCGAUGUGCGGGGCUAAAGAGCUAGGAGAAA ..---........(((.....(((((((((.(((((........((.....)).....))))).....)))).)))))...)))..((((....))))........ ( -26.62) >DroSec_CAF1 32649 97 + 1 AAAAAAAAAGGGGGUAACAAGCGGUGGGUC-------GUGGGGAGCGGCGAGCGGAGGACCUGGAGUGGACCGCACCGAUGUGCGGGGCUAAAGAGCUAGG--AAA .................(...(.((..(((-------((.....)))))..)).)..).(((((..(((.((((((....))))))..))).....)))))--... ( -28.00) >DroYak_CAF1 32400 87 + 1 AA---AAGGGGGGGUAACUAGCAGGGGGUGGCGGGAA--------------GCGGAGGACCUGGAGUGGACCGCAUCGCUGUGCGGGGCUAAAGAGCUAGG--AAA ..---............((((((((...(.((.....--------------)).)....)))....(((.((((((....))))))..)))....))))).--... ( -23.90) >consensus AA___AAAAGGGGGUAACAAGCGGUGGGUC_CGGG_A_UGGG_AGCGGC_AGCGGAGGACCUGGAGUGGACCGCACCGAUGUGCGGGGCUAAAGAGCUAGG__AAA ...........................................................(((((..(((.((((((....))))))..))).....)))))..... (-18.15 = -17.93 + -0.22)

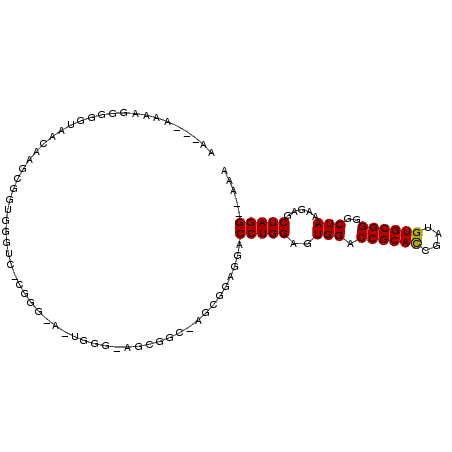
| Location | 11,418,177 – 11,418,280 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11418177 103 - 20766785 UUUCUCCUAGCUCUUUAGCCCCGCACAUCGGUGCGGUCCACUCCAGGUCCUCCGCUUGCCGCUACCCAUUACCCGUAACCCACCGCUUGUUACCCCCUUUA---UU .......((((......(..((((((....))))))..)....(((((.....)))))..))))..........(((((.........)))))........---.. ( -19.50) >DroSec_CAF1 32649 97 - 1 UUU--CCUAGCUCUUUAGCCCCGCACAUCGGUGCGGUCCACUCCAGGUCCUCCGCUCGCCGCUCCCCAC-------GACCCACCGCUUGUUACCCCCUUUUUUUUU ...--....(((....))).....(((.(((((.((((.......((....))((.....)).......-------)))))))))..)))................ ( -18.70) >DroYak_CAF1 32400 87 - 1 UUU--CCUAGCUCUUUAGCCCCGCACAGCGAUGCGGUCCACUCCAGGUCCUCCGC--------------UUCCCGCCACCCCCUGCUAGUUACCCCCCCUU---UU ...--.(((((......((...))...(((..((((.((......))....))))--------------....)))........)))))............---.. ( -17.10) >consensus UUU__CCUAGCUCUUUAGCCCCGCACAUCGGUGCGGUCCACUCCAGGUCCUCCGCU_GCCGCU_CCCA_U_CCCG_AACCCACCGCUUGUUACCCCCUUUU___UU ........(((......(..((((((....))))))..)......((....))...............................)))................... (-12.97 = -13.30 + 0.33)

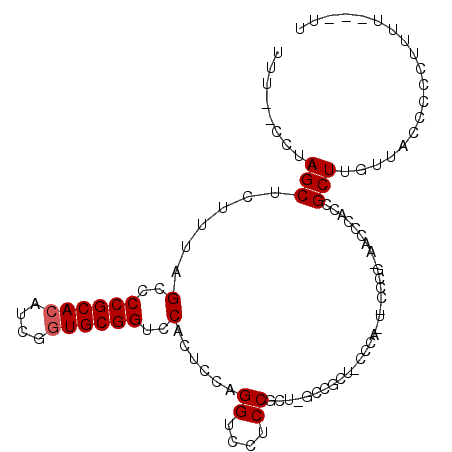
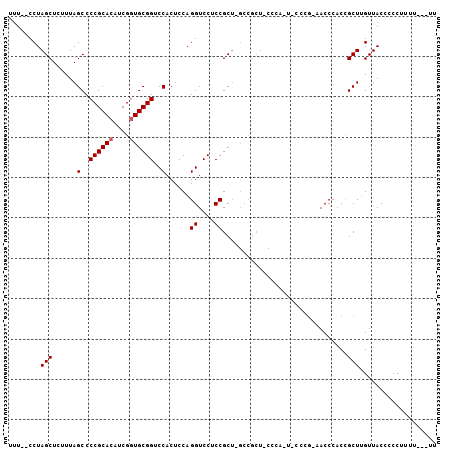
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:54 2006