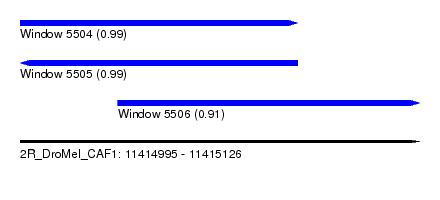
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,414,995 – 11,415,126 |
| Length | 131 |
| Max. P | 0.990052 |

| Location | 11,414,995 – 11,415,086 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -19.98 |
| Energy contribution | -21.35 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11414995 91 + 20766785 AUGUCUAAAA--------AAAAAUAUAUCUGUUGGUGCUGACUGGUGACCCCAUUUGUGGGGUGACCCUGCCAUGUGGUGACACCUUAACAGUUAGCCA ..........--------................(.((((((((((.((((((....)))))).))........(((....))).....))))))))). ( -30.10) >DroSec_CAF1 29540 88 + 1 AUGUCUAAAA--------GAA---AUAUCUGUUGGUGCUGGCUGGUGACCCCAUUUGUGGGGUGACCCCGCCAUGUGGUGACACCUUAACAGUUAGCCA ..........--------...---.((.(((((((...((((.(((.((((((....)))))).)))..)))).(((....))).))))))).)).... ( -32.10) >DroEre_CAF1 30760 78 + 1 AUGCCUUUAA--------AAA---AUAUCUGUUG----------GUGACCCCAUUUGUGGGGUGACCCUGCCAUGUGGAGACACCUUUACUGUUAGCCA ..........--------...---.........(----------((.((((((....)))))).)))(((.((.((((((....)))))))).)))... ( -22.00) >DroYak_CAF1 29320 92 + 1 AUGCCUUUAAAUGUCUUAAAA---AUAUUUGUUGGUGCU----GGUGACCCCAUUUAUGGGGUGACCCUGCCAUGUGGAGACACCUUUACAGUUAGCCA ..(((..(((((((.......---)))))))..)))(((----(((.((((((....)))))).)))(((....(((....))).....)))..))).. ( -30.70) >consensus AUGCCUAAAA________AAA___AUAUCUGUUGGUGCU____GGUGACCCCAUUUGUGGGGUGACCCUGCCAUGUGGAGACACCUUAACAGUUAGCCA .........................((.((((.((((......(((.((((((....)))))).))).((((....)))).))))...)))).)).... (-19.98 = -21.35 + 1.37)

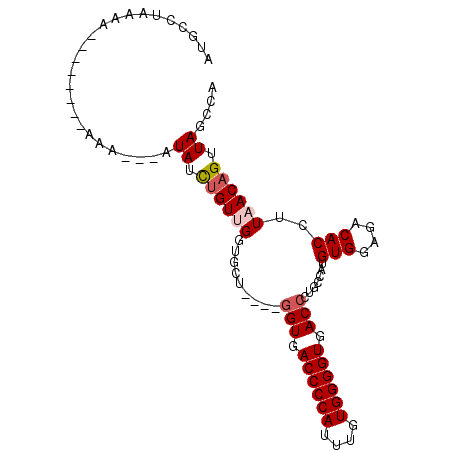
| Location | 11,414,995 – 11,415,086 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -18.35 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11414995 91 - 20766785 UGGCUAACUGUUAAGGUGUCACCACAUGGCAGGGUCACCCCACAAAUGGGGUCACCAGUCAGCACCAACAGAUAUAUUUUU--------UUUUAGACAU .......(((((..(((((.......((((..(((.((((((....)))))).))).))))))))))))))..........--------.......... ( -27.11) >DroSec_CAF1 29540 88 - 1 UGGCUAACUGUUAAGGUGUCACCACAUGGCGGGGUCACCCCACAAAUGGGGUCACCAGCCAGCACCAACAGAUAU---UUC--------UUUUAGACAU .......(((((..(((((.......((((..(((.((((((....)))))).))).))))))))))))))....---...--------.......... ( -31.01) >DroEre_CAF1 30760 78 - 1 UGGCUAACAGUAAAGGUGUCUCCACAUGGCAGGGUCACCCCACAAAUGGGGUCAC----------CAACAGAUAU---UUU--------UUAAAGGCAU ..(((......((((((((((((....))...(((.((((((....)))))).))----------)...))))))---)))--------)....))).. ( -23.70) >DroYak_CAF1 29320 92 - 1 UGGCUAACUGUAAAGGUGUCUCCACAUGGCAGGGUCACCCCAUAAAUGGGGUCACC----AGCACCAACAAAUAU---UUUUAAGACAUUUAAAGGCAU ..(((.........(((((((((....)).))(((.((((((....)))))).)))----.))))).........---.((((((...))))))))).. ( -25.30) >consensus UGGCUAACUGUAAAGGUGUCACCACAUGGCAGGGUCACCCCACAAAUGGGGUCACC____AGCACCAACAGAUAU___UUU________UUAAAGACAU ((.((..(((((...(((....)))..)))))(((.((((((....)))))).))).....................................)).)). (-18.35 = -19.23 + 0.87)

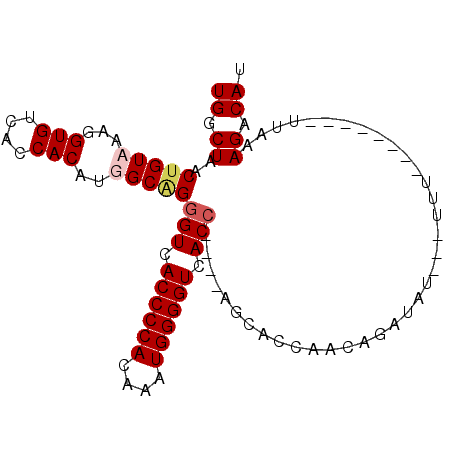
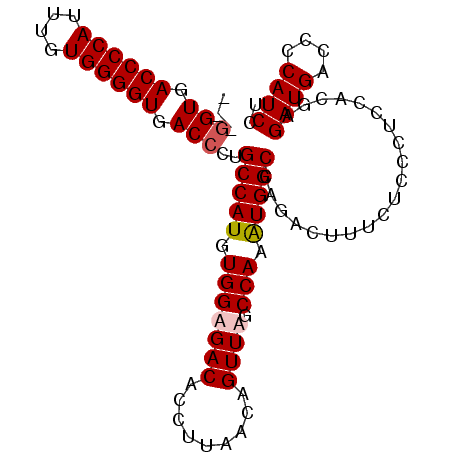
| Location | 11,415,027 – 11,415,126 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -27.77 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11415027 99 + 20766785 ACUGGUGACCCCAUUUGUGGGGUGACCCUGCCAUGUGGUGACACCUUAACAGUUAGCCAAGUGGCCAGACUAUUUCCCUCCACGAGAUGACCCCAUCCU ...(((.((((((....)))))).)))(((((((.(((((((.........)))).))).)))).))).................((((....)))).. ( -32.10) >DroSec_CAF1 29569 98 + 1 GCUGGUGACCCCAUUUGUGGGGUGACCCCGCCAUGUGGUGACACCUUAACAGUUAGCCAAGUGGCCAGACU-UUUCCCUCCACGAGAUGACCCCAUCCU ...(((.((((((....)))))).)))..(((((.(((((((.........)))).))).)))))......-.............((((....)))).. ( -31.40) >DroEre_CAF1 30783 95 + 1 ----GUGACCCCAUUUGUGGGGUGACCCUGCCAUGUGGAGACACCUUUACUGUUAGCCAAAUGGCCAGACUUUCUCCCCCCAUGAGAUGACCCCAUCCU ----((.((((((....)))))).)).(((((((.(((.((((.......))))..))).)))).))).................((((....)))).. ( -28.00) >DroYak_CAF1 29356 96 + 1 ---GGUGACCCCAUUUAUGGGGUGACCCUGCCAUGUGGAGACACCUUUACAGUUAGCCAAAUGGCCAGACUUUCUCCCACCAUGAGAUGACCCCAUCCU ---(((.((((((....)))))).)))....((((.(((((.........((((.(((....)))..)))).)))))...)))).((((....)))).. ( -31.00) >consensus ___GGUGACCCCAUUUGUGGGGUGACCCUGCCAUGUGGAGACACCUUAACAGUUAGCCAAAUGGCCAGACUUUCUCCCUCCACGAGAUGACCCCAUCCU ...(((.((((((....)))))).)))..(((((.(((((((.........)))).))).)))))....................((((....)))).. (-27.77 = -28.27 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:51 2006