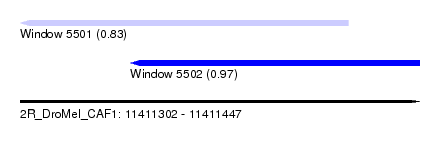
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,411,302 – 11,411,447 |
| Length | 145 |
| Max. P | 0.970736 |

| Location | 11,411,302 – 11,411,421 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -18.06 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.833943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11411302 119 - 20766785 UACAAAGCUAAGCCUUUUGCUUACUUUGUGGCUAGCCAUGCAAUUGGCCAGCAAAGUGAGUAGAUAUGCACUUAAAUCAUGCCUAAAAUCAGUCUUUUAACAGAGGAUAACUUACAUUU (((((((.(((((.....))))))))))))(((.((((......)))).)))...((((((.(((..(((.........))).....))).((((((.....)))))).)))))).... ( -31.50) >DroSec_CAF1 24349 107 - 1 UACAAA------GCUUUUGCUUACUUUCUGGCUAACCAUGCAAUUUGUCAGCAAAGUAAGUAGAUAUGCACUUAAAUCAACACUAAAU--GGU-AUUUAUGAG---AAAACUUACAUUU ......------((.(((((((((((((((((..............))))).))))))))))))...))......((((........)--)))-.....((((---....))))..... ( -21.44) >DroSim_CAF1 23977 116 - 1 UACAAAGCUAAAGCUUUUGCUUACUUUCUGGCUAACCAUGCAAUUGGUCAGCAAAGUAAGUAGAUAUGAACUUAAAUCAGUACUAAAU--GGU-AUUUAUCAGAUGAAAACUUACAUUU ...(((((....)))))((((((((((((((((((........)))))))).))))))))))(((((((.......)))(((((....--)))-)).))))(((((........))))) ( -27.10) >DroEre_CAF1 27032 115 - 1 UACUAGGCUAAAGCUUUUGCUUACUUUCUGGCUAGCCAUGCAAUUGGUCAGCAGAGUAAGUAGAUAUGCACUUAAUUCAGCCAUAAAC--GUC-UUUUGUCACAGGUAAACUUAC-UUU ((((.((((.((((.((((((((((((((((((((........)))))))).))))))))))))...))......)).))))......--(.(-....).)...)))).......-... ( -28.20) >DroYak_CAF1 25526 116 - 1 UACAAAUCUAAAGUUAAUACUUACUUUCUGGAUAGCCACGCAAUUGGUCAGCAGAGUACGUAGAUGAGCACUUAAAUCAGCCCUAAAU--GUC-UUUUGUCAGAGGUAAAAAUACAUUU ............(((..(((.(((((((((....((((......))))))).)))))).)))....)))............(((...(--(.(-....).)).)))............. ( -17.70) >consensus UACAAAGCUAAAGCUUUUGCUUACUUUCUGGCUAGCCAUGCAAUUGGUCAGCAAAGUAAGUAGAUAUGCACUUAAAUCAGCCCUAAAU__GGU_UUUUAUCAGAGGAAAACUUACAUUU ............((.((((((((((((((((((((........)))))))).))))))))))))...)).................................................. (-18.06 = -18.54 + 0.48)


| Location | 11,411,342 – 11,411,447 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -22.19 |
| Energy contribution | -23.27 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11411342 105 - 20766785 CGUGAUCACCGAUGACUUAGGAUGUAUACAAAGCUAAGCCUUUUGCUUACUUUGUGGCUAGCCAUGCAAUUGGCCAGCAAAGUGAGUAGAUAUGCACUUAAAUCA ............(((.(((((.((((((....((...))..((((((((((((((((((((........))))))).)))))))))))))))))))))))).))) ( -32.00) >DroSec_CAF1 24383 99 - 1 CGUGAUCACCGAUGAUUUAGGAUGUAUACAAA------GCUUUUGCUUACUUUCUGGCUAACCAUGCAAUUUGUCAGCAAAGUAAGUAGAUAUGCACUUAAAUCA ............(((((((((.((((((....------...(((((((((((((((((..............))))).))))))))))))))))))))))))))) ( -29.75) >DroSim_CAF1 24014 105 - 1 CGUGAUCACCGAUGAUUUAGGAUGUAUACAAAGCUAAAGCUUUUGCUUACUUUCUGGCUAACCAUGCAAUUGGUCAGCAAAGUAAGUAGAUAUGAACUUAAAUCA ............(((((((((...((((((((((....)))))((((((((((((((((((........)))))))).))))))))))).))))..))))))))) ( -31.40) >DroEre_CAF1 27068 105 - 1 CGUGAUCGCCGAUGCCUUAGGAUGUAUACUAGGCUAAAGCUUUUGCUUACUUUCUGGCUAGCCAUGCAAUUGGUCAGCAGAGUAAGUAGAUAUGCACUUAAUUCA .(((...((....((((.((........))))))....)).((((((((((((((((((((........)))))))).))))))))))))....)))........ ( -32.20) >DroYak_CAF1 25563 105 - 1 UGUGAUCACCGAUGCCGUAGGAUGUAUACAAAUCUAAAGUUAAUACUUACUUUCUGGAUAGCCACGCAAUUGGUCAGCAGAGUACGUAGAUGAGCACUUAAAUCA .(((.(((.(.........((((........))))........(((.(((((((((....((((......))))))).)))))).)))).))).)))........ ( -18.90) >consensus CGUGAUCACCGAUGACUUAGGAUGUAUACAAAGCUAAAGCUUUUGCUUACUUUCUGGCUAGCCAUGCAAUUGGUCAGCAAAGUAAGUAGAUAUGCACUUAAAUCA ............(((.(((((.((((((.............((((((((((((((((((((........)))))))).))))))))))))))))))))))).))) (-22.19 = -23.27 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:47 2006