| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,393,168 – 11,393,281 |
| Length | 113 |
| Max. P | 0.939340 |

| Location | 11,393,168 – 11,393,281 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -17.27 |
| Energy contribution | -16.50 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
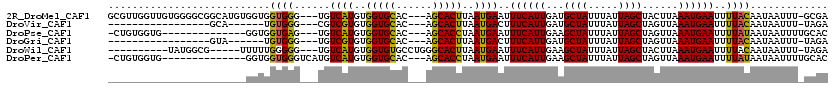
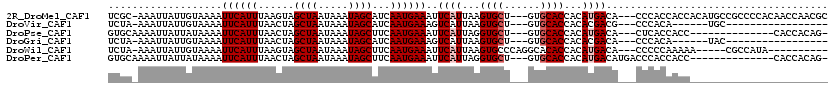
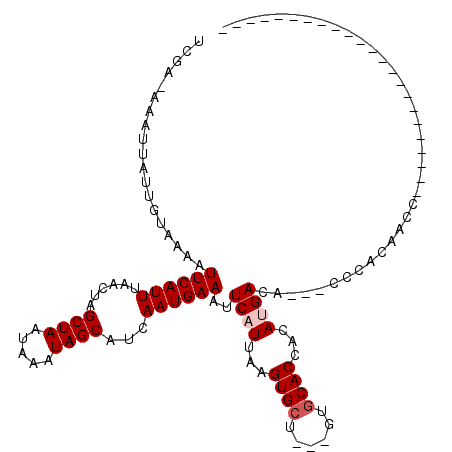
>2R_DroMel_CAF1 11393168 113 + 20766785 GCGUUGGUUGUGGGGCGGCAUGUGGUGGUGGG---UGUCAUGUGGUGCAC---AGCACUUAAUGAAUUUCAUUGAUGCUAUUUAUUAGCUACUUAAAUGAAUUUUACAAUAAUUU-GCGA (((...(((((((((...(((..((((((..(---..((....))..)..---((((.((((((.....))))))))))........))))))...)))..)))))))))....)-)).. ( -29.70) >DroVir_CAF1 12231 90 + 1 -----------------GCA------UGUGGG---CGUCGUGUGGUGCAC---AGCACUUAAUGACUUUCAUUGAUGCUAUUUAUUAGCUAGUUAAAUGAAUUUUACAAUAAUUU-UAGA -----------------...------((((((---.(((((..(((((..---.)))))..))))).((((((...((((.....))))......)))))).)))))).......-.... ( -20.00) >DroPse_CAF1 9936 99 + 1 -CUGUGGUG--------------GGUGGUGAG---UGUCAUGUGGUGCAC---AGCACCUAAUGAAUUUCAUUGAAGCUAUUUAUUAGCUAGUUAAAUGAAUUUUAUAAUAAUUUUGCAC -.((..(((--------------((((.((.(---..((....))..).)---).))))))(((((.((((((..(((((.....))))).....)))))).))))).......)..)). ( -28.30) >DroGri_CAF1 11442 90 + 1 -----------------GUA------UGUGGG---UGUCGUGUGGUGCAC---AGCACUUAAUGACUUUCAUUGAUGCUAUUUAUUAGCUAGUUAAAUGAAUUUUACAAUAAUUU-UAGA -----------------...------((((((---.(((((..(((((..---.)))))..))))).((((((...((((.....))))......)))))).)))))).......-.... ( -19.80) >DroWil_CAF1 12150 101 + 1 ----------UAUGGCG-----UUUUUGGGGG---UGUCAUGUGGUGUGCCUGGGCACUUAAUGAAUUUCAUUGAAGCUAUUUAUUAGCUACUUAAAUGAAUUUUACAAUAAUUU-UAGA ----------(((((((-----..(....)..---)))))))....((((....))))....((((.((((((..(((((.....))))).....)))))).)))).........-.... ( -22.10) >DroPer_CAF1 10938 102 + 1 -CUGUGGUG--------------GGUGGUGGGUCAUGUCAUGUGGUGCAC---AGCACCUAAUGAAUUUCAUUGAAGCUAUUUAUUAGCUAGUUAAAUGAAUUUUAUAAUAAUUUUGCAC -.((..(((--------------(((((((.(((((.....))))).)))---..))))))(((((.((((((..(((((.....))))).....)))))).))))).......)..)). ( -25.70) >consensus _______________________GGUGGUGGG___UGUCAUGUGGUGCAC___AGCACUUAAUGAAUUUCAUUGAAGCUAUUUAUUAGCUAGUUAAAUGAAUUUUACAAUAAUUU_GAGA ...........................((((......((((..(((((......)))))..))))..((((((...((((.....))))......))))))..))))............. (-17.27 = -16.50 + -0.77)
| Location | 11,393,168 – 11,393,281 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -14.30 |
| Consensus MFE | -9.83 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
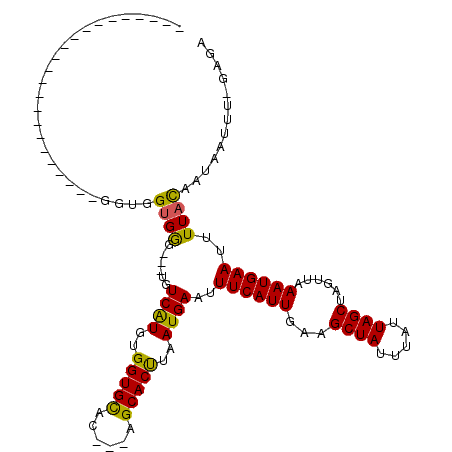
>2R_DroMel_CAF1 11393168 113 - 20766785 UCGC-AAAUUAUUGUAAAAUUCAUUUAAGUAGCUAAUAAAUAGCAUCAAUGAAAUUCAUUAAGUGCU---GUGCACCACAUGACA---CCCACCACCACAUGCCGCCCCACAACCAACGC ..((-(.............((((((...((.((((.....)))))).))))))..((((...((((.---..))))...))))..---............)))................. ( -12.80) >DroVir_CAF1 12231 90 - 1 UCUA-AAAUUAUUGUAAAAUUCAUUUAACUAGCUAAUAAAUAGCAUCAAUGAAAGUCAUUAAGUGCU---GUGCACCACACGACG---CCCACA------UGC----------------- ....-.......(((....((((((......((((.....))))...)))))).(((.....((((.---..)))).....))).---...)))------...----------------- ( -11.40) >DroPse_CAF1 9936 99 - 1 GUGCAAAAUUAUUAUAAAAUUCAUUUAACUAGCUAAUAAAUAGCUUCAAUGAAAUUCAUUAGGUGCU---GUGCACCACAUGACA---CUCACCACC--------------CACCACAG- (((................((((((.....(((((.....)))))..))))))..((((..(((((.---..)))))..))))..---.........--------------...)))..- ( -17.80) >DroGri_CAF1 11442 90 - 1 UCUA-AAAUUAUUGUAAAAUUCAUUUAACUAGCUAAUAAAUAGCAUCAAUGAAAGUCAUUAAGUGCU---GUGCACCACACGACA---CCCACA------UAC----------------- ....-.......(((....((((((......((((.....))))...)))))).(((.....((((.---..)))).....))).---...)))------...----------------- ( -11.40) >DroWil_CAF1 12150 101 - 1 UCUA-AAAUUAUUGUAAAAUUCAUUUAAGUAGCUAAUAAAUAGCUUCAAUGAAAUUCAUUAAGUGCCCAGGCACACCACAUGACA---CCCCCAAAAA-----CGCCAUA---------- ....-.......(((....((((((.....(((((.....)))))..)))))).........((((....))))...))).....---..........-----.......---------- ( -14.60) >DroPer_CAF1 10938 102 - 1 GUGCAAAAUUAUUAUAAAAUUCAUUUAACUAGCUAAUAAAUAGCUUCAAUGAAAUUCAUUAGGUGCU---GUGCACCACAUGACAUGACCCACCACC--------------CACCACAG- (((................((((((.....(((((.....)))))..))))))..((((..(((((.---..)))))..))))..............--------------...)))..- ( -17.80) >consensus UCGA_AAAUUAUUGUAAAAUUCAUUUAACUAGCUAAUAAAUAGCAUCAAUGAAAUUCAUUAAGUGCU___GUGCACCACAUGACA___CCCACAACC_______________________ ...................((((((......((((.....))))...))))))..((((...((((......))))...))))..................................... ( -9.83 = -10.33 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:40 2006